XB-IMG-126525
Xenbase Image ID: 126525
|
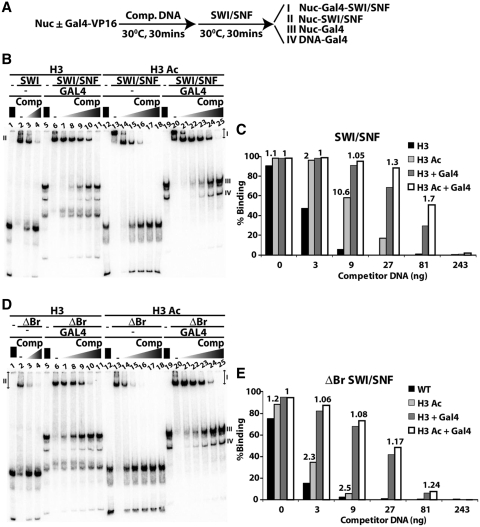
Figure 4. SWI/SNF recruitment by the transcription activator Gal4-VP16 and H3 acetylation. (A) The order of addition of nucleosomes, competitor DNA and SWI/SNF is shown for the recruitment assays in (B and D). Nucleosomes contain one Gal4 site in extranucleosomal DNA, 27 bp from the entry site. Gel shift assays are shown with 15 nM SWI/SNF (B) or 10 nm ΔBr SWI/SNF (D) that had 10 nM nucleosomes and 25 nM Gal4-VP16 where indicated. H3 acetylated nucleosomes (H3 Ac) are in lanes 12–25 and nucleosomes without acetylation (H3) are in lanes 1–11. Increasing amounts of competitor DNA were added ranging from 0.45 to 35 ng/µl for lanes 7–11 and 21–25 or from 0.45 to 1.3 ng/µl for lanes 3–4 and 14–18. Species I, II, III and IV refer to nucleosome-Gal4-SWI/SNF, nucleosome-SWI/SNF, nucleosome-Gal4 and DNA-Gal4, respectively. Quantification of (B and D) are shown in (C and E), respectively, for SWI/SNF and ΔBr SWI/SNF binding to H3 Ac versus H3 nucleosomes with and without Gal4-VP16. The numbers above the bars is the ratio of H3 Ac versus H3 nucleosomes binding with or without Gal4-VP16 added. The binding ratios are included for those particular gel shift lanes in which each species could be accurately quantified. Image published in: Chatterjee N et al. (2011) © The Author(s) 2011. This image is reproduced with permission of the journal and the copyright holder. This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
