XB-IMG-138159
Xenbase Image ID: 138159
|
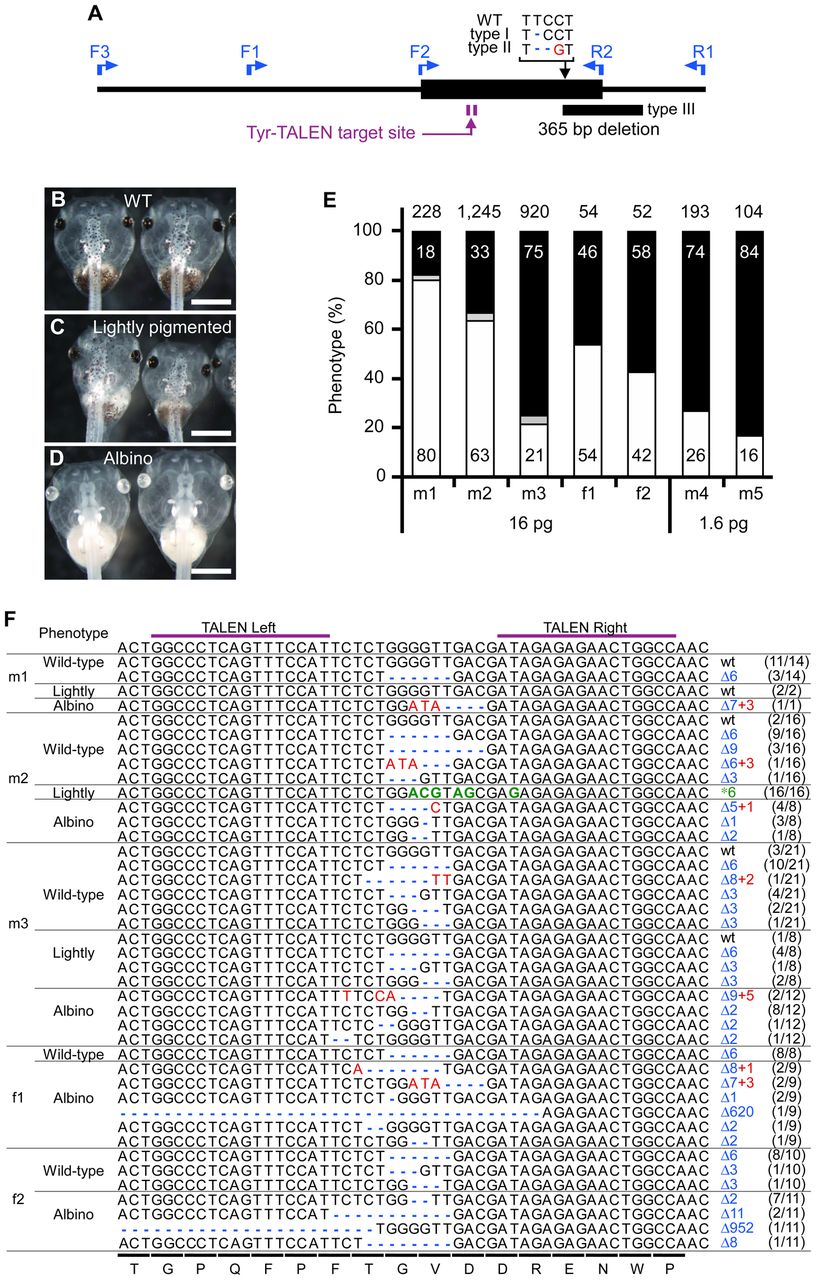
Fig. 3. Analysis of individual offspring obtained from crosses between F0 and albino frogs.
(A) Schematic representation showing both the gene mutations of the albino mates used in the crosses and the locations of primers used in the mutation analysis. The bold horizontal line indicates the DNA region from the initiation codon to the end of the first exon. The mutated sequences of the albino mates are shown as type I, type II and type III with the wild-type sequence. The inserted and deleted nucleotides are indicated with a red character and blue dashes, respectively. The locations of the mutations and the 365-bp deletion in the albino mates are indicated by a black arrow and rectangle, respectively. The Tyr-TALEN target site and the direction of the primers are indicated by a purple arrow and blue arrows, respectively. (B–D) Brightfield images of live wild-type (B), lightly pigmented (C) and albino (D) tadpoles. Note that the abdominal region is lightly pigmented in C. Scale bars = 1 mm. (E) Phenotypic analysis of the offspring of F0 and albino frogs. The percentages of wild-type, lightly pigmented and albino F1 tadpoles are indicated by black, gray and white bars, respectively (Table 1). The numbers in the bars are the percentages of tadpoles with the indicated phenotype. The total number of tadpoles is shown at the top of the bars. (F) Genotypic analysis of individual offspring of F0 and albino frogs. The target DNA fragment was amplified using genomic DNA purified from individual wild-type, lightly pigmented and albino F1 tadpoles and was recloned for sequence determination. All of the mating data are shown. Different sequences were bequeathed by F0 frogs to different F1 offspring, which also had the other mutated tyrosinase allele inherited from the albino parent. The target sequences derived from the albino mates are not shown. The wild-type target DNA and the amino acid sequences are indicated at the top and bottom of the panel, respectively. A pair of purple bars denotes the TALEN-binding sites. The gaps resulting from a deletion (Δ), the inserted nucleotides (+) and the exchanged nucleotides (*) are indicated with blue dashes, red characters and green characters, respectively. The mutation types are indicated on the right. The ratio of the number of tadpoles with the indicated sequence to the total number of tadpoles with the same phenotype is shown in parentheses on the right. Large deletions of 620 and 952 nucleotides were observed in the offspring derived from the f1 and f2 frogs, respectively, and both of these deletions contained the start codon. Image published in: Nakajima K and Yaoita Y (2015) © 2015. Creative Commons Attribution license Larger Image Printer Friendly View |
