XB-IMG-147736
Xenbase Image ID: 147736
|
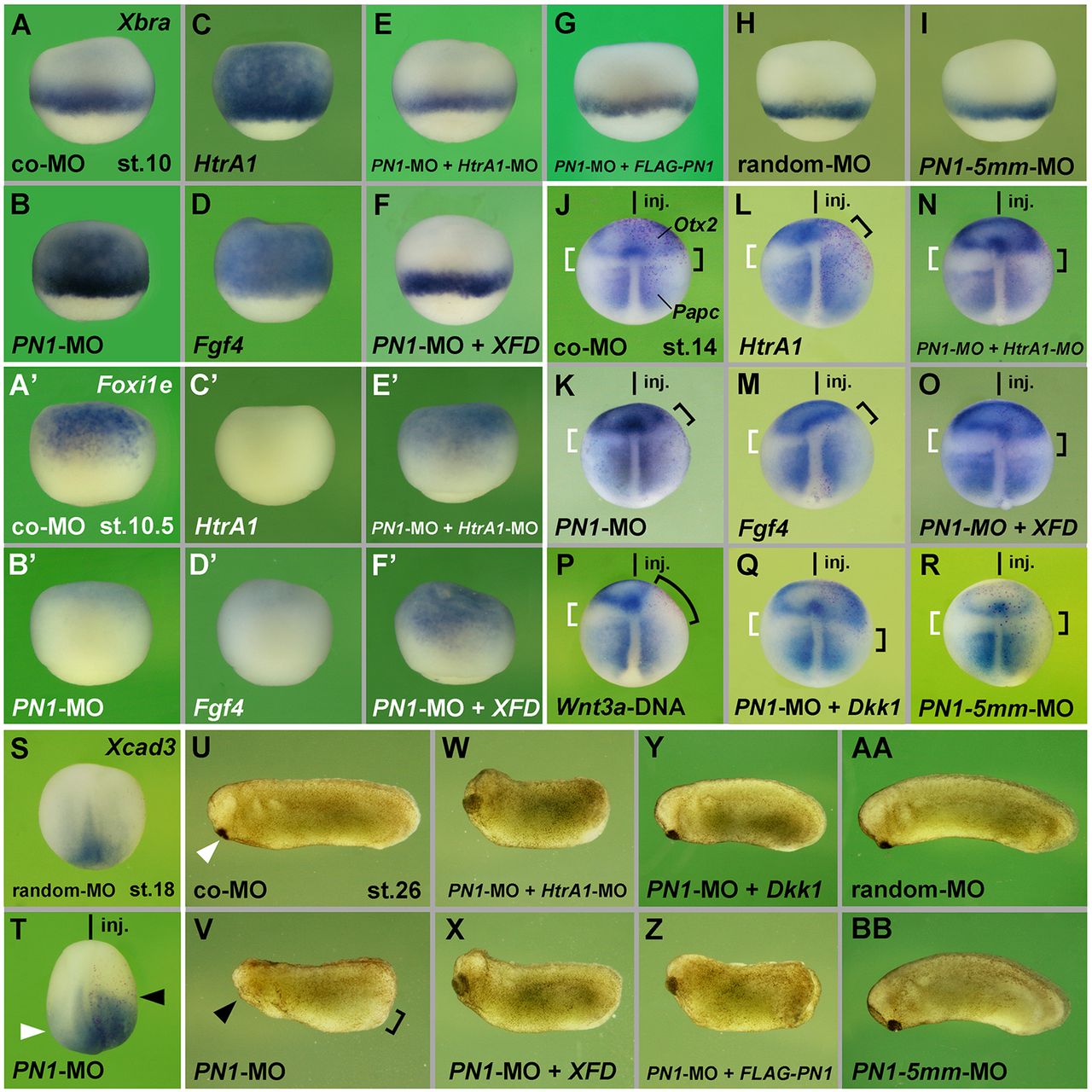
Fig. 5.
PN1 functions in an HtrA1-, FGF- and Wnt-dependent manner. (A-Dâ²) Lateral view of early gastrula embryos. PN1-MO, HtrA1 mRNA and Fgf4 mRNA, but not co-MO, induce ectopic Xbra expression (A-D) and a reduction of Foxi1e expression (Aâ²-Dâ²) in the animal hemisphere. (E-Fâ²) In PN1 morphant embryos, 20â
ng HtrA1-MO and XFD mRNA restore normal expression of Xbra and Foxi1e. (G-I) Random-MO, PN1-5mm-MO, and a combination of PN1-MO and non-targeted FLAG-PN1 mRNA do not affect Xbra expression. (J-T) Dorsal view of neurulae. A single injection of PN1-MO shifts the border between Otx2 and Papc (brackets in K) and expands Xcad3 expression (arrowheads in T) anteriorward. Co-MO and PN1-5mm-MO have no effect on Otx2 and Papc (J,R), and random-MO has no effect on Xcad3 expression (S). HtrA1 mRNA, Fgf4 mRNA and Wnt3a-DNA reduce Otx2, but only HtrA1 and Fgf4 expand Papc expression (L,M,P). Co-injections of 5â
ng HtrA1-MO, XFD and Dkk1 mRNA revert the effect of PN1-MO and cause slight expansion of Otx2 and posteriorward retraction of Papc signals (N,O,Q). (U-BB) PN1-MO reduces head structures (arrowhead) and expands the proctodeum (bracket in V), whereas co-MO, random-MO and PN1-5mm-MO have no effect in tailbud embryos (U,AA,BB). 20â
ng HtrA1-MO, XFD, Dkk1 and FLAG-PN1 rescue posteriorization in PN1 morphants (W-Z). Injected mRNA amounts per embryo were: HtrA1, 200â
pg (50â
pg in L); Fgf4, 2â
pg (0.5â
pg in M); XFD, 80â
pg (20â
pg in O); FLAG-PN1, 800â
pg; Dkk1, 24â
pg (8â
pg in Q). Indicated phenotypes were shown by: A, 136/144; Aâ², 50/59; B, 154/186; Bâ², 50/69; C, 21/21; Câ², 47/57; D, 79/79; Dâ², 93/97; E, 66/90; Eâ², 52/56; F, 72/83; Fâ², 45/51; G, 24/30; H, 44/46; I, 14/24; J, 12/13; K, 14/14; L, 60/60; M, 45/49; N, 17/17; O, 9/9; P, 34/36; Q, 55/55; R, 31/32; S, 8/8; T, 28/30; U, 75/77; V, 97/95; W, 48/59; X, 23/24; Y, 67/65; Z, 22/23; AA, 7/10; BB, 10/15. Image published in: Acosta H et al. (2015) Copyright © 2015. Image reproduced with permission of the Publisher.
Image source: Published Larger Image Printer Friendly View |
