XB-IMG-156027
Xenbase Image ID: 156027
|
|
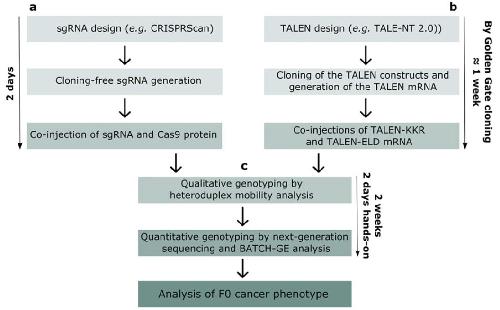
FIGURE 1 Flowchart demonstrating our pipe-line for generation of GEXM by CRISPR/Cas9 (left) or TALEN techniques (right), converging
at the employed methods for assessing genome editing efficiencies in F0 mosaic GEXM (center). (Top left) sgRNA is designed using the
CRISPRScan online tool, and subsequently synthesized by a cloning-free method (Moreno-Mateos et al., 2015). For this, two DNA oligonucleotides
are annealed and extended in a PCR reaction and the resulting dsDNA template is used for in vitro transcription generating
injection-ready sgRNA, which is subsequently coinjected with Cas9 protein (Nakayama et al., 2014; Top right) TALEN pairs are designed
using TALE-NT 2.0 online tool, and are generated by Golden Gate cloning (Cermak et al., 2015; Doyle et al., 2012). Synthetic mRNAs
encoding obligate heterodimeric TALEN-KKR and TALEN-ELD pairs are subsequently co-injected. (Center) Independent of the choice of targeted
nuclease, HMA is used for qualitative genotyping of the F0 mosaic GEXM. If genome editing can be detected, qualitative genotyping
is performed by targeted deep next-generation sequencing and BATCH-GE analysis (Boel et al., 2016) Image published in: Naert T et al. (2017) Copyright © 2017. Image reproduced with permission of the Publisher, John Wiley & Sons. Larger Image Printer Friendly View |
