XB-IMG-169497
Xenbase Image ID: 169497
|
|
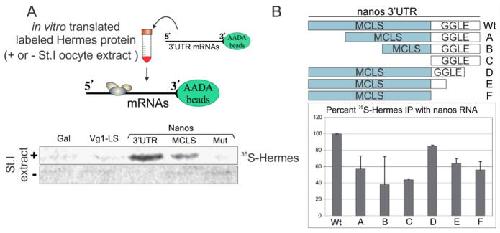
Figure 2. Hermes/Rbpms specifically associates with nanos RNA in a UGCAC dependent manner. (A) Hermes/Rbpms interacts with nanos but not Vg1 RNA. Vg1 3′UTR Localization Experimental design is shown at top. Vg1 Localization Signal (LS), nanos1 3′UTR, nanos Mitochondrial Cloud Localization Signal (MCLS), and a nanos mutant MCLS (mut) were tested for Hermes/Rbpms binding. Individual RNAs were immobilized on AADA agarose beads and mixed with in vitro translated Hermes/Rbpms protein labeled with 35S-methionine. Bound proteins were analyzed by SDS-PAGE and autoradiography. Galactosidase (Gal) RNA served as a negative control. Hermes/Rbpms only bound the nanos1–3′UTR and only when reactions included stage I/II oocyte extracts and not reticulocyte lysates alone. Note: the 3′UTR binds more Hermes/Rbpms than the MCLS alone suggesting the GGLE may also bind Hermes/Rbpms; (B) Both the MCLS and GGLE are required for wild-type levels of Hermes/Rbpms binding and require stage I oocyte extract. The deletion mutants diagramed were immobilized on AADA beads and analyzed as described in Methods. Error bars indicate standard deviation of three experiments. Hermes/Rbpms binding to the full-length nanos 3′UTR is set at 100%. Note: both regions contribute to Hermes binding. Image published in: Aguero T et al. (2016) Image reproduced on Xenbase with permission of the publisher and the copyright holder. This image is reproduced with permission of the journal and the copyright holder. This is an open-access article distributed under the terms of the Creative Commons Attribution license Larger Image Printer Friendly View |
