XB-IMG-170123
Xenbase Image ID: 170123
|
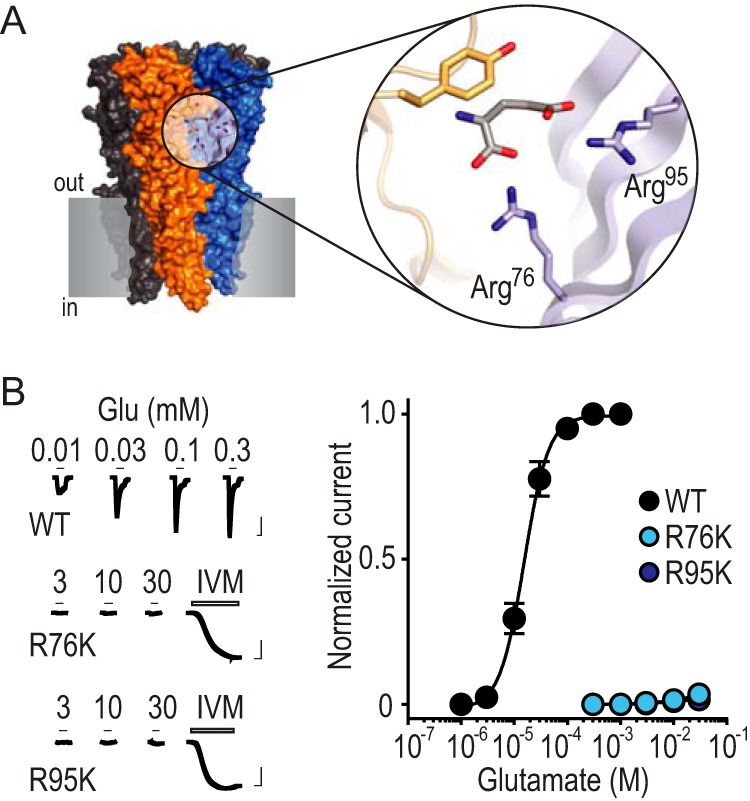
FIGURE 1. Arg-Lys substitutions drastically reduce glutamate recognition.
A, X-ray structure of GLC-1 GluCl (PDB 3RIF; gray shading, notional cell membrane). Magnified view shows glutamate binding site and selected amino acid side chains. These include GLC-1 arginine residues 37 and 56, which are labeled Arg76 and Arg95 to describe the equivalent residues from the AVR-14B GluCl used in the present study. B, left, example recordings of glutamate (Glu) and ivermectin (IVM, 1 μm) responses at oocytes expressing mutant AVR-14B GluCls (scale bars: x, 5 s; y, 2 μA). Activation by IVM, which binds elsewhere on the receptor, confirms cell surface expression in the absence of Glu-gated currents. Right, mean ± S.E. (n = 4–8) peak current responses to increasing concentrations of Glu, normalized to maximum Glu-gated current (WT) or maximum IVM-gated current (mutants). Image published in: Lynagh T et al. (2017) © 2017 by The American Society for Biochemistry and Molecular Biology, Inc. This image is reproduced with permission of the journal and the copyright holder. This is an open-access article distributed under the terms of the Creative Commons Attribution license Larger Image Printer Friendly View |
