XB-IMG-173294
Xenbase Image ID: 173294
|
Fig. S5. Preference of cyclin B/Cdk and cyclinA/Cdk for C/sites of Otx2.
(A) Schematic presentation of Myc@Otx2ΔAD with double (2A) and triple alanine (3A)
mutations at S116, S132 and S158. To test a preference of cyclin B/Cdk and cyclin
A/Cdks for Otx2 phosphorylation, double alanine mutants (2A) for S116, S132 and
S158 in ΔAD were constructed, in which only one site is responsible for Otx2
phosphorylation. In the 2A constructs, for example, the S158 construct indicates
mutations at S116 and S132. Yellow box, Myc@tagY black box, homeodomain (HD)Y
blue box, repression domain (RD). S, serine phosphorylation sites (S116, S132,
S158)Y A, alanine mutation. (B) Western blotting of Myc@Otx2ΔAD constructs (2A or
3A) co@expressed with (+) or without (@) HA@cyclin B1*. The intensity of modified
bands of S132 (S116A/S158A) and S116 (S132A/S158A) constructs, but not that of
S158 (S116A/S132A), was increased by HA@cyclin B1* expression. (C) Western
blotting of Myc@Otx2ΔAD constructs (2A or 3A) co@expressed with (+) or without (@)
HA@cyclin A1*. All modified bands were enhanced by HA@cyclin A1* expression, with
an additional modified band (white arrowheads). These data suggest that S116 and
S132 have a preference for both cyclin B/Cdk and cyclin A/Cdks, and that S158 has it
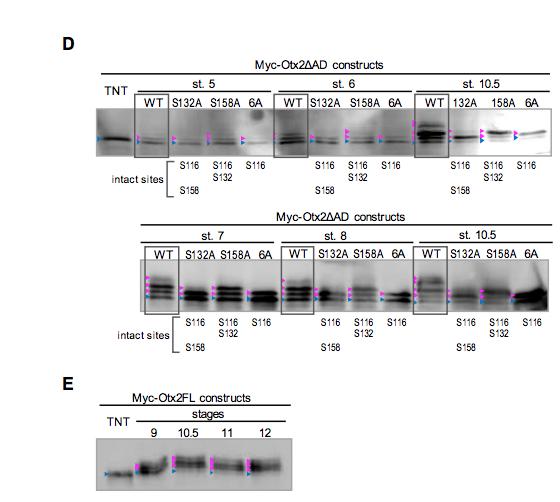
for cyclin A/Cdks, if Otx2 is directly phosphorylated by cyclin/Cdks. (D,E)
Developmental changes of phosphorylation of Otx2 as assayed by using Myc@
Otx2ΔAD (D) and Myc@Otx2FL (E) constructs. The wild type (WT) and alanine
mutants of a single serine at S132 (S132A) and S158 (S158A), and of 6 serines at
S122, S123, S132, S153, S158, and S161 (6A) in Myc@Otx2ΔAD constructs were
analyzed from stages 5 to 10.5 as indicated. Note that the 6A construct has S116,
which is a phosphorylation site. Mutation at S132 (S132A and 6A constructs)
reduced phosphorylation levels compared to S158 mutation, indicating that S132 and
S116 are more efficiently phosphorylated than S158 during cleavage stages. Myc@
Otx2FL was analyzed at stages 9 to 12 as indicated. TNT, in vitro translation
productsY blue arrowheads, nascent bandsY magenta arrowheads, modified bands.
Blue arrowheads, nascent proteinsY magenta arrowheads, modified proteinsY white
arrowheads, additional modified bands at sites other than S116, S132 and S158. Image published in: Satou Y et al. (2018) Copyright © 2018. Image reproduced with permission of the publisher and the copyright holder. This is an Open Access article distributed under the terms of the Creative Commons Attribution License. Larger Image Printer Friendly View |
