XB-IMG-75343
Xenbase Image ID: 75343
|
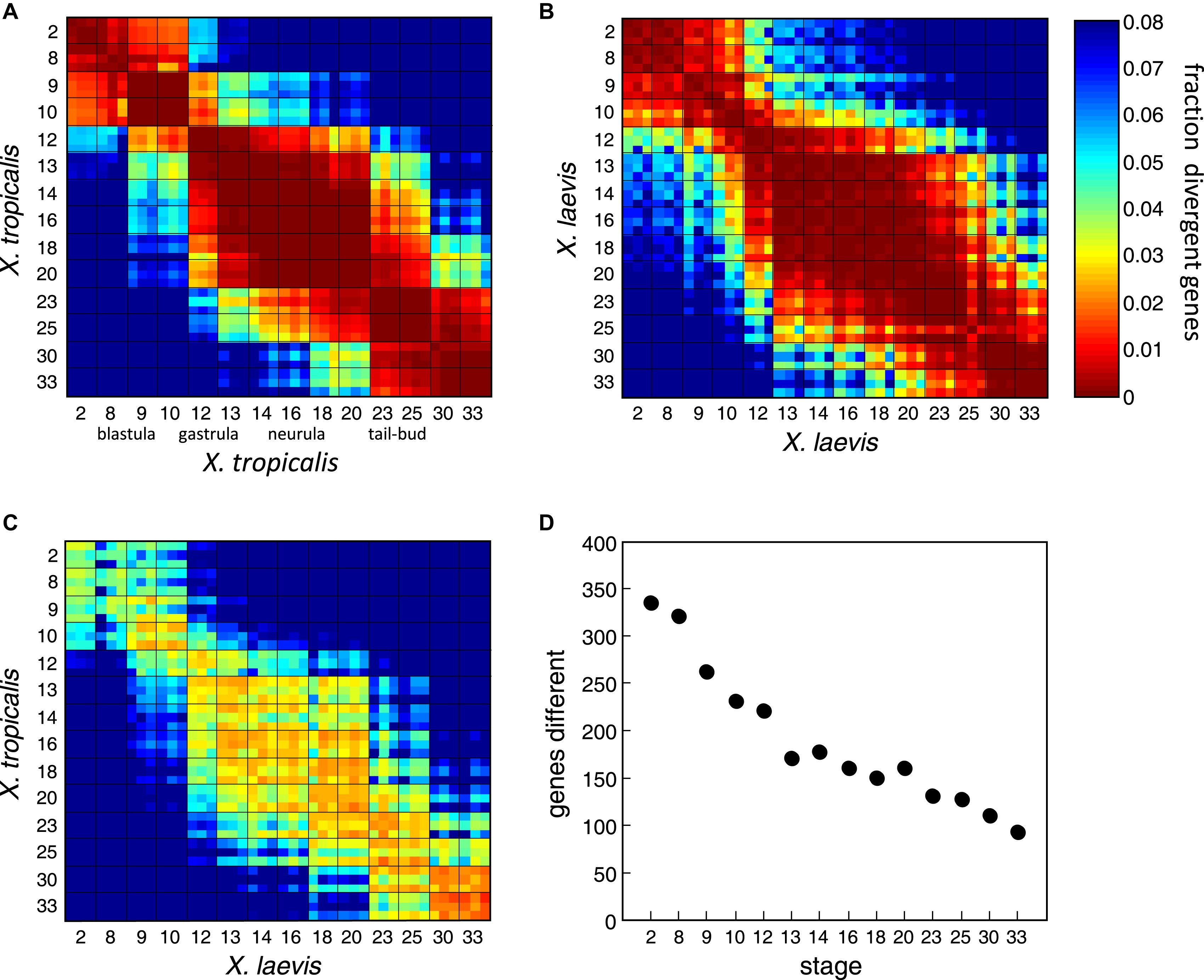
Figure 5. Global Comparison of the X. laevis and X. tropicalis Developmental Transcriptomes(AâC) The heat maps represent the fraction of genes significantly different between pairs of transcriptomes; the grid separates the replicates across the stages. The color of each square indicates the difference at the specified developmental stages. (A) and (B) represent X. tropicalis and X. laevis plotted against themselves.(C) X. tropicalis plotted against X. laevis. The diagonal squares by definition have zero divergence. The fraction is computed as the number of genes with a difference of at least 1.5 log10 units out of the number of genes with a maximum expression of at least 2.5 log10 units in either transcriptomes.(D) For 2297 dynamically expressed genes, the plot indicates the number of genes with significantly different transcript abundance (>1.5 log10). Image published in: Yanai I et al. (2011) Copyright © 2011. Image reproduced with permission of the Publisher, Elsevier B. V. Larger Image Printer Friendly View |
