XB-IMG-77924
Xenbase Image ID: 77924
|
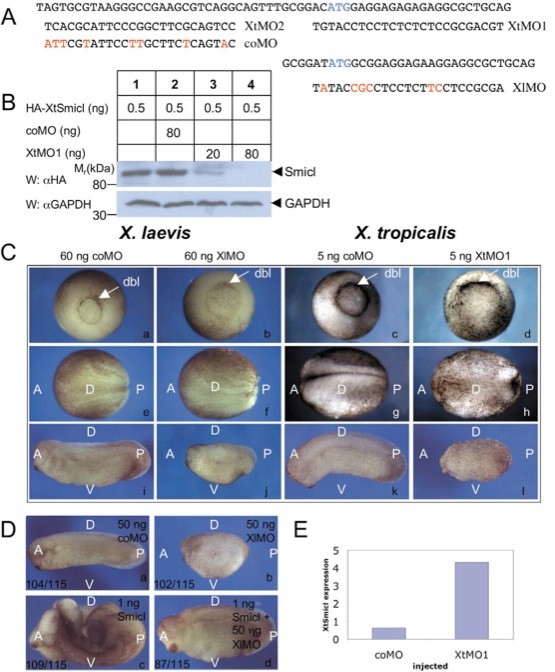
Fig. 2. Analysis of Smicl function in Xenopus laevis and Xenopus tropicalis. (A) The antisense morpholino oligonucleotides used in this study are aligned with their Xenopus tropicalis (Xt) and Xenopus laevis (Xl) target sequences. (B) XtMO1 inhibits translation of RNA encoding HA-tagged XtSmicl in a dose-dependent fashion. This is not observed with coMO. XtMO1 and coMO were injected in Xenopus embryos at the one-cell stage at the indicated concentrations, followed by RNA encoding HA-tagged XtSmicl. Embryos were cultured to early gastrula stage 10 and subjected to western blotting using an anti-HA antibody and an anti-Gapdh antibody as a loading control. (C) Injection of the morpholino oligonucleotides described in A disrupts gastrulation and axis formation in both Xenopus laevis and Xenopus tropicalis. Dbl, dorsal blastopore lip; A, anterior; P, posterior; D, dorsal; V, ventral. (D) Injection of mRNA encoding mouse Smicl into embryos of Xenopus laevis can escuethe phenotype caused by XlMO. Overexpression of mouse Smicl alone causes a pina bifidaphenotype. Quantitation of morpholino defects and rescues are indicated on the figure. (E) Injection of antisense morpholino oligonucleotide XtMO1 causes upregulation of Smicl mRNA. RNA was extracted at stage 10.5 from embryos injected with 5 ng XtMO1 or coMO and analysed by quantitative RT-PCR. Image published in: Collart C et al. (2005) Copyright © 2005. Image reproduced with permission of the Publisher and the copyright holder. This is an Open Access article distributed under the terms of the Creative Commons Attribution License. Larger Image Printer Friendly View |
