Searchable Databases of Xenopus RNA-Seq Expression Profiles
In addition to external RNA-seq databases like the SRA at NCBI and gene expression data at GEO, a growing number of Xenopus laboratories are generating high-resolution RNA-seq datasets of Xenopus gene transcription during development. Highlighted resources are as follows. The publication titles in the table below link to each database. For more details, please see below for highlights and links to each publications featuring these valuable resources.
______________
| Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. | Owens et al., Cell Rep. January 26, 2016; 14 (3): 632-47. | XB-ART-51804 | ||
| On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development. | Peshkin et al., Dev Cell. November 9, 2015; 35 (3): 383-94. | XB-ART-51556 | ||
| High-resolution analysis of gene activity during the Xenopus mid-blastula transition. | Collart et al., Development. May 1, 2014; 141 (9): 1927-39. | XB-ART-48872 | ||
| RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. | Tan et al., Genome Res. January 1, 2013; 23 (1): 201-16. | XB-ART-45933 |
______________
Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development.
Owens ND , Blitz IL , Lane MA , Patrushev I , Overton JD , Gilchrist MJ , Cho KW , Khokha MK.
Cell Rep. January 26, 2016; 14 (3): 632-47.
Transcript regulation is essential for cell function, and misregulation can lead to disease. Despite tech- nologies to survey the transcriptome, we lack a comprehensive understanding of transcript kinetics, which limits quantitative biology. This is an acute challenge in embryonic development, where rapid changes in gene expression dictate cell fate deci- sions. By ultra-high-frequency sampling of Xenopus embryos and absolute normalization of sequence reads, we present smooth gene expression trajec- tories in absolute transcript numbers. During a devel- opmental period approximating the first 8 weeks of human gestation, transcript kinetics vary by eight orders of magnitude. Ordering genes by expression dynamics, we find that ‘‘temporal synexpression’’ predicts common gene function. Remarkably, a single parameter, the characteristic timescale, can classify transcript kinetics globally and distinguish genes regulating development from those involved in cellular metabolism. Overall, our analysis provides unprecedented insight into the reorganization of maternal and embryonic transcripts and redefines our ability to perform quantitative biology.
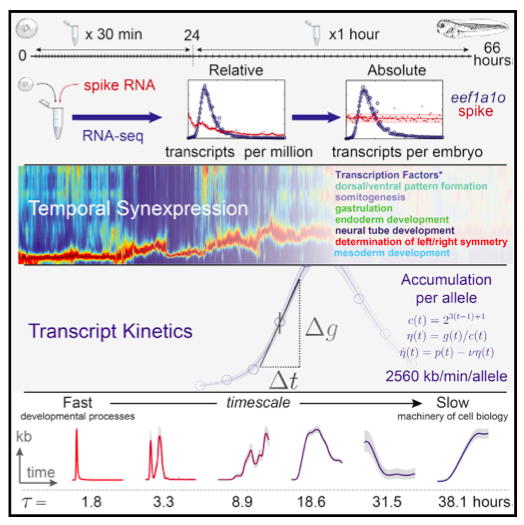
Follow this link to find the searchable database of gene transcript levels from RNA-seq data generated by the Gilchrist lab, Cho lab, and Khokha lab.
Searching this database displays gene expression levels from 0 to 66 hours post-fertilization in X. tropicalis.
Search by gene or retrieve lists of genes whose expression levels are activated at a selected certain time point.
Transcription factor and signaling molecule genes can be filtered from search results.
On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development.
Peshkin L , Wühr M , Pearl E , Haas W , Freeman RM , Gerhart JC , Klein AM , Horb M , Gygi SP , Kirschner MW.
Dev Cell. November 9, 2015; 35 (3): 383-94.
Summary
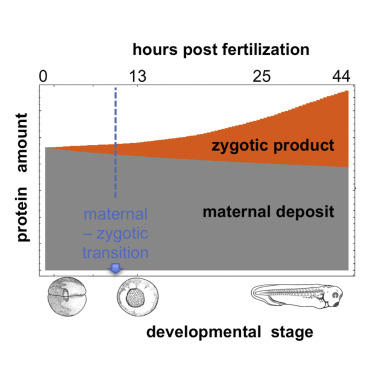
A biochemical explanation of development from the fertilized egg to the adult requires an understanding of the proteins and RNAs expressed over time during embryogenesis. We present a comprehensive characterization of protein and mRNA dynamics across early development in Xenopus. Surprisingly, we find that most protein levels change little and duplicated genes are expressed similarly. While the correlation between protein and mRNA levels is poor, a mass action kinetics model parameterized using protein synthesis and degradation rates regresses protein dynamics to RNA dynamics, corrected for initial protein concentration. This study provides detailed data for absolute levels of ∼10,000 proteins and ∼28,000 transcripts via a convenient web portal, a rich resource for developmental biologists. It underscores the lasting impact of maternal dowry, finds surprisingly few cases where degradation alone drives a change in protein level, and highlights the importance of transcription in shaping the dynamics of the embryonic proteome.
Follow this link: http://kirschner.med.harvard.edu/MADX.html to query the data from the Kirshner lab.
This server provides supplementary tables and browser for "On the relationship of protein and mRNA dynamics in vertebrate embryonic development", Peshkin et al., Developmental Cell, 2015.
The paper can be accessed free through Dec. 29, 2015 by following this link: http://authors.elsevier.com/a/1S0Dp5Sx5gPQfn
Or, can be accessed from Developmental Cell here: http://www.sciencedirect.com/science/article/pii/S1534580715006577
High-resolution analysis of gene activity during the Xenopus mid-blastula transition.
Collart C , Owens ND , Bhaw-Rosun L , Cooper B , De Domenico E , Patrushev I , Sesay AK , Smith JN , Smith JC , Gilchrist MJ.
Development. May 1, 2014; 141 (9): 1927-39.
The Xenopus mid-blastula transition (MBT) marks the onset of large-scale zygotic transcription, as well as an increase in cell cycle length and a loss of synchronous cell divisions. Little is known about what triggers the activation of transcription or how newly expressed genes interact with each other. Here, we use high-resolution expression profiling to identify three waves of gene activity: a post-fertilisation wave involving polyadenylation of maternal transcripts; a broad wave of zygotic transcription detectable as early as the seventh cleavage and extending beyond the MBT at the twelfth cleavage; and a shorter post-MBT wave of transcription that becomes apparent as development proceeds. Our studies have also allowed us to define a set of maternal mRNAs that are deadenylated shortly after fertilisation, and are likely to be degraded thereafter. Experimental analysis indicates that the polyadenylation of maternal transcripts is necessary for the establishment of proper levels of zygotic transcription at the MBT, and that genes activated in the second wave of expression, including Brachyury and Mixer, contribute to the regulation of genes expressed in the third. Together, our high-resolution time series and experimental studies have yielded a deeper understanding of the temporal organisation of gene regulatory networks in the early Xenopus embryo.
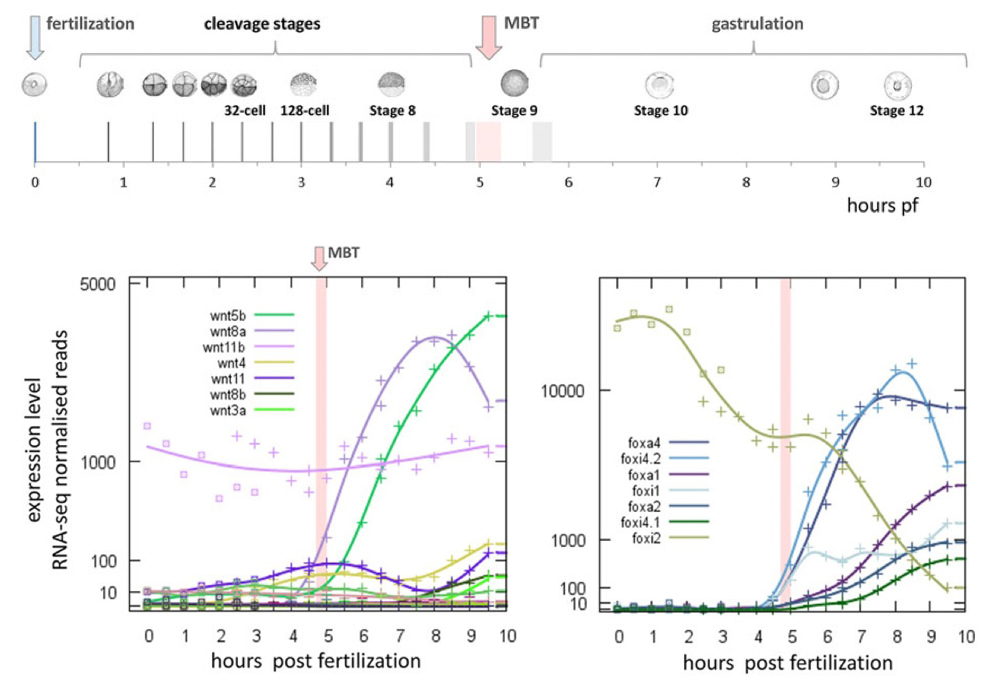
Follow this link to find the searchable database of gene transcript levels from RNA-seq data generated by the Gilchrist lab.
Searching this database displays gene expression levels from 0 to 10 hours post-fertilization in X. tropicalis.
Fertilization at 0 hours, mid-blastula transition at approximately 5 hours, to the end of gastrulation at 10 hours.
Search by gene or retrieve lists of genes whose expression levels are activated at a selected certain time point.
Transcription factor and signaling molecule genes can be filtered from search results.
Click here to see the Xenbase literature page.
Click here to view literature details from PubMed.
Click to view this publication in Development or PubMed Central.
RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development.
Tan MH , Au KF , Yablonovitch AL , Wills AE , Chuang J , Baker JC , Wong WH , Li JB.
Genome Res. January 1, 2013; 23 (1): 201-16.
The Xenopus embryo has provided key insights into fate specification, the cell cycle, and other fundamental developmental and cellular processes, yet a comprehensive understanding of its transcriptome is lacking. Here, we used paired end RNA sequencing (RNA-seq) to explore the transcriptome of Xenopus tropicalis in 23 distinct developmental stages. We determined expression levels of all genes annotated in RefSeq and Ensembl and showed for the first time on a genome-wide scale that, despite a general state of transcriptional silence in the earliest stages of development, approximately 150 genes are transcribed prior to the midblastula transition. In addition, our splicing analysis uncovered more than 10,000 novel splice junctions at each stage and revealed that many known genes have additional unannotated isoforms. Furthermore, we used Cufflinks to reconstruct transcripts from our RNA-seq data and found that ∼13.5% of the final contigs are derived from novel transcribed regions, both within introns and in intergenic regions. We then developed a filtering pipeline to separate protein-coding transcripts from noncoding RNAs and identified a confident set of 6686 noncoding transcripts in 3859 genomic loci. Since the current reference genome, XenTro3, consists of hundreds of scaffolds instead of full chromosomes, we also performed de novo reconstruction of the transcriptome using Trinity and uncovered hundreds of transcripts that are missing from the genome. Collectively, our data will not only aid in completing the assembly of the Xenopus tropicalis genome but will also serve as a valuable resource for gene discovery and for unraveling the fundamental mechanisms of vertebrate embryogenesis.
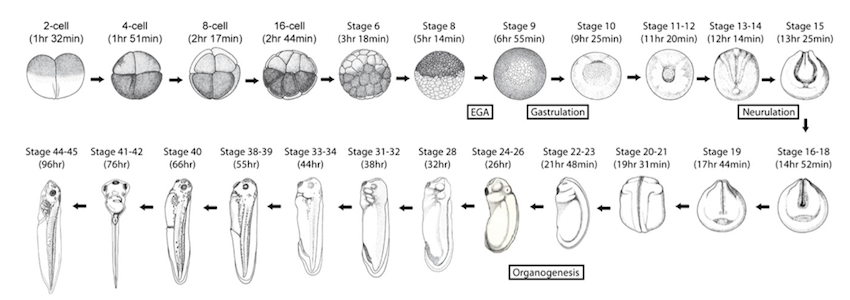
Follow this link to see the visualization of these transcriptome datasets.
This searchable database spans the 2-cell embryo through tadpole stages.
Query through RefSeq or Ensembl IDs.
Click here to see the Xenbase literature page.
Click here to view literature details from PubMed.
Click to view this publication in Genome Research or PubMed Central.
