De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome (BAMS) and abrogate nasal development
Gordon et al.
Nat Genet. January 9, 2017
View article at Nature Genetics
View article on Pubmed
View article on Xenbase
Abstract
Bosma arhinia microphthalmia syndrome (BAMS) is an extremely rare and striking condition characterized by complete absence of the nose with or without ocular defects. We report here that missense mutations in the epigenetic regulator SMCHD1 mapping to the extended ATPase domain of the encoded protein cause BAMS in all 14 cases studied. All mutations were de novo where parental DNA was available. Biochemical tests and in vivo assays in Xenopus laevis embryos suggest that these mutations may behave as gain-of-function alleles. This finding is in contrast to the loss-of-function mutations in SMCHD1 that have been associated with facioscapulohumeral muscular dystrophy (FSHD) type 2. Our results establish SMCHD1 as a key player in nasal development and provide biochemical insight into its enzymatic function that may be exploited for development of therapeutics for FSHD.
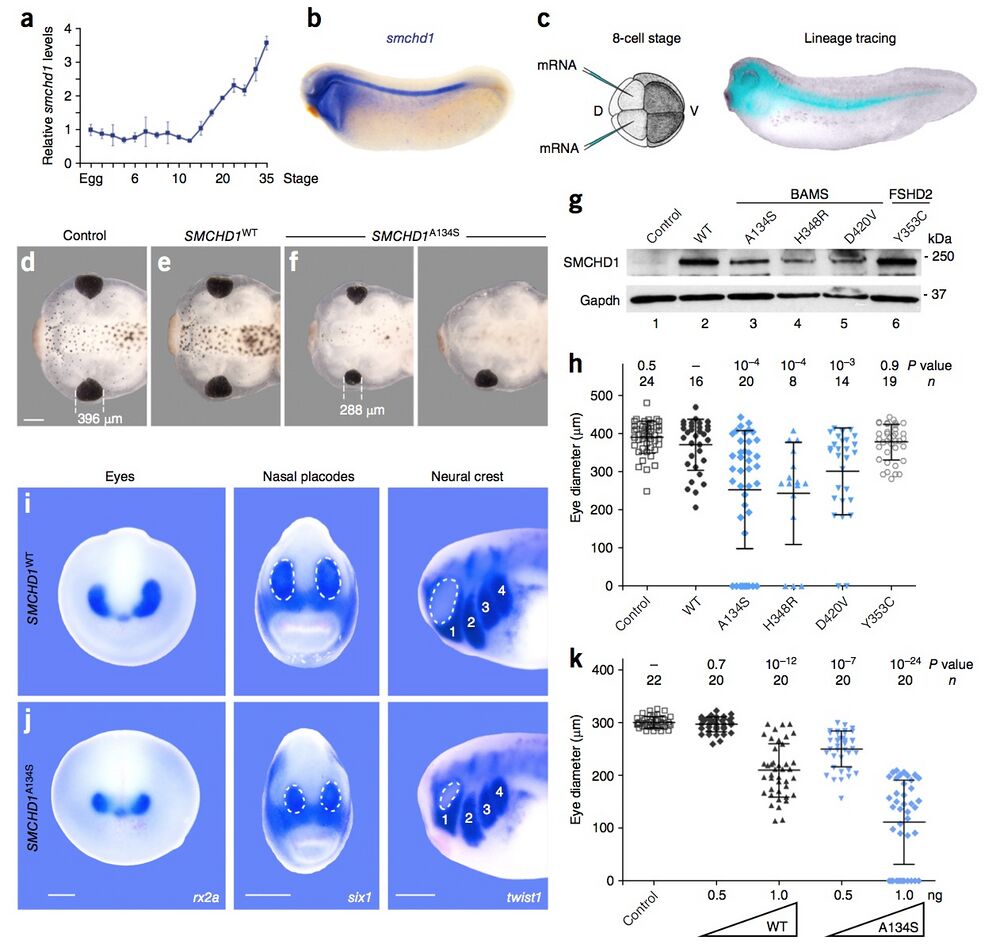
Figure 3: In vivo functional assays in Xenopus embryos suggest that BAMS-associated mutations behave as gain-of-function alleles. (a) Expression of smchd1 relative to 18S rRNA by qPCR. Data represent means ± s.d. of triplicates. (b) In late tailbud stages, smchd1 expression is restricted to the head region and the neural tube. (c) To target the head structures, dorsal–animal blastomeres in eight-cell-stage Xenopus embryos were injected with synthesized mRNA (120 pg for all panels except k). These cells are fated to give rise to head structures, as shown by dextran lineage tracing. D, dorsal; V, ventral. (d–f) Representative stage 45 tadpoles that are uninjected (d) or injected with SMCHD1WT (e) or SMCHD1A134S (f) mRNA. Those injected with SMCHD1A134S mRNA display craniofacial anomalies and smaller eyes in comparison to control tadpoles and those injected with SMCHD1WT mRNA. Scale bar, 0.3 mm. All images were acquired at the same magnification. (g) Immunoblot of stage 12 embryonic extracts from control and injected embryos showing expression of exogenous human SMCHD1. (h) Eye diameter is significantly reduced in embryos overexpressing BAMS- associated mutants (blue) relative to siblings overexpressing wild-type SMCHD1 (black) or embryos overexpressing an FSHD2-associated mutant (open circles). (i,j) In situ hybridization for rx2a, six1 and twist1, demarcating the eyes, placodes and neural crest, respectively, in embryos injected with SMCHD1WT (i) or SMCHD1A134S (j) mRNA. Images are representative of 9 of 10, 7 of 10, and 10 of 10 embryos for each probe. The dotted lines outline nasal placodes (middle) and the eye (right). Numbers label streams of migrating cranial neural crest. Scale bars, 0.2 mm (same magnification for each comparison of i to j). (k) Measurements of eye diameter for Xenopus embryos injected with 0.5 or 1 ng of mRNA encoding wild-type SMCHD1 or a BAMS-associated mutant show that SMCHD1 overexpression causes dose-dependent craniofacial anomalies. Biological variation between clutches of tadpoles is seen in the data presented in h and k. For both h and k, n indicates the number of embryos analyzed, data are shown as means ± s.d. and
P values were calculated by Kruskal–Wallis test followed by Dunn’s post test.
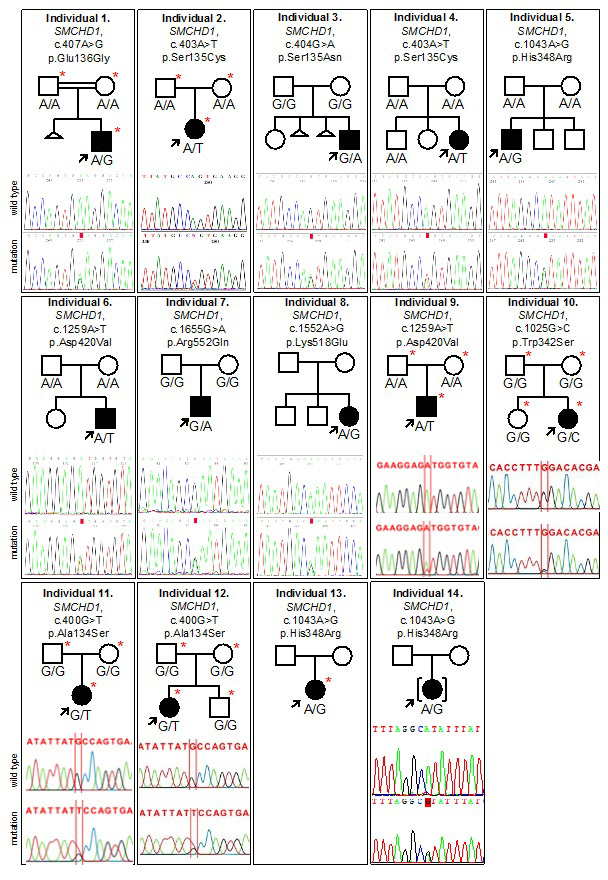
Supplementary Figure 2: BAMS pedigrees and Sanger sequencing chromatograms of SMCHD1 mutations. Individuals submitted for exome sequencing are indicated by a red asterisk. Note that Sanger sequencing was unavailable for individual 13.
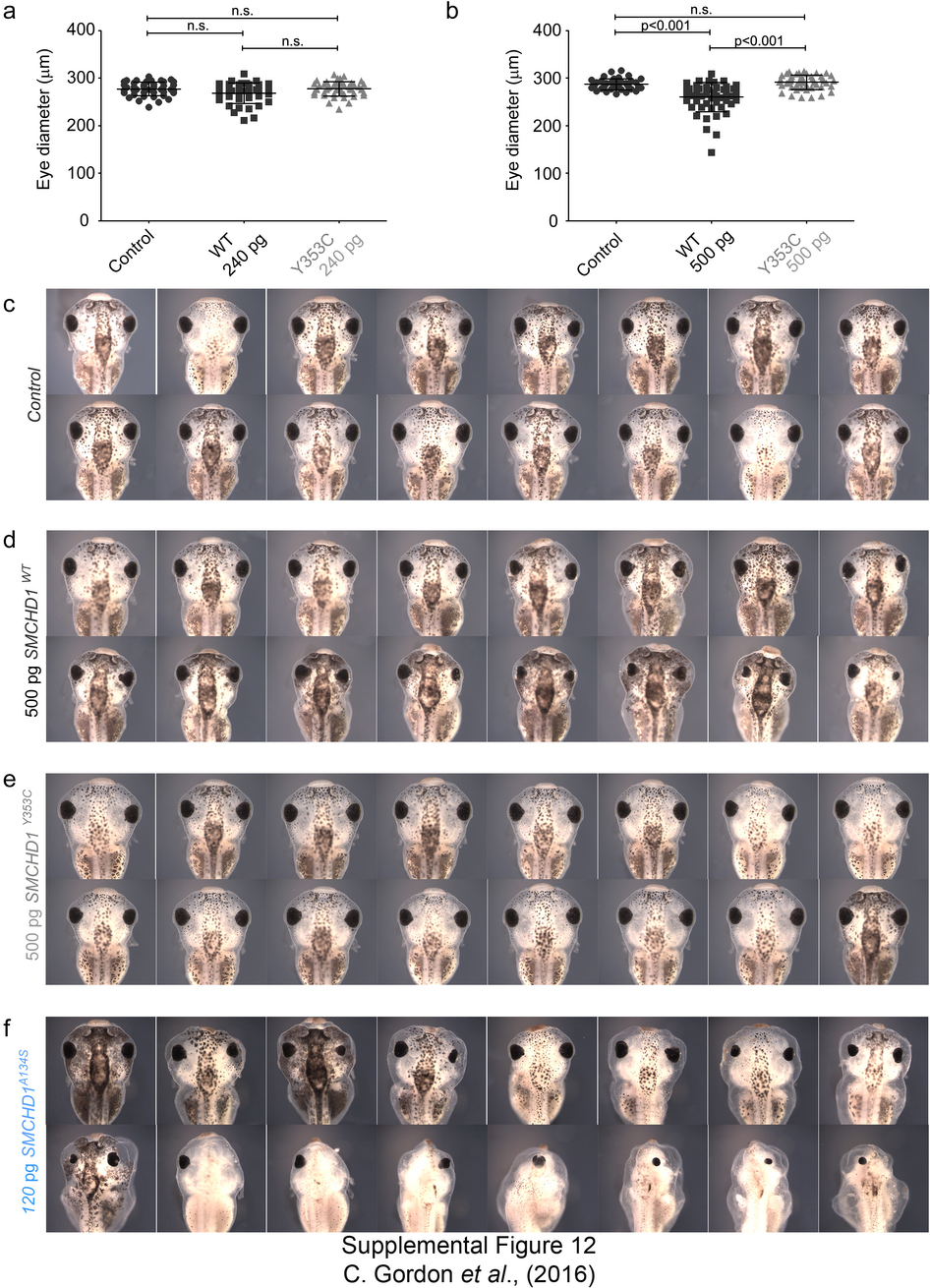
Supplementary Figure 12: SMCHD1 overexpression in Xenopus causes dose-dependent craniofacial anomalies. (a,b) Measurements of eye diameter of Xenopus embryos injected with 240 pg (a) or 500 pg (b) SMCHD1 mRNA. Y353C is an FSHD2 mutation. At least 20 embryos were studied for each condition. (c–f) Representative Xenopus embryos injected with 500 pg of wild-type or FSHD2 mutant SMCHD1 or 120 pg of BAMS mutant mRNA show varying degrees of craniofacial abnormalities as compared to uninjected control tadpoles at 4 days post-fertilization. Data are shown as means ± s.d. P values were calculated by Kruskal–Wallis test followed by Dunn’s post test. n.s., not significant.
Adapted with permission from Macmillan Publishers Ltd: Gordon et al. (2017).Spinal cord regeneration in Xenopus laevis. Nature Genetics (2017) doi:10.1038/ng.3765, copyright (2017).
