Autism risk gene dyrk1a is required for ciliogenesis and brain size
Summary
Willsey & Xu, et al., identify novel molecular functions for the neurodevelopmental disorder risk gene DYRK1A during vertebrate development. Patients with mutations in DYRK1A often have autism, microcephaly, intellectual disability, and kidney defects. These researchers used Xenopus tropicalis to discover that mutations in DYRK1A cause defects in cell cycle progression and ciliogenesis. These disruptions provide insights about disorder etiology, as disruption in cell division can lead to microcephaly and disruption of ciliogenesis often leading to kidney and/or heart defects. This work also has important implications for Down Syndrome and Alzheimer’s Disease, as DYRK1A inhibitors are currently being tested as therapeutics.
Click here to view article at Development.
Click here to view article on Pubmed.
Click here to view article on Xenbase.
Neurodevelopmental disorder risk gene DYRK1A is required for ciliogenesis and brain size in Xenopus embryos
Helen Rankin Willsey, Yuxiao Xu, Amanda Everitt, Jeanselle Dea, Cameron R. T. Exner, A. Jeremy Willsey, Matthew W. State, Richard M. Harland
Development 2020 : dev.189290 doi: 10.1242/dev.189290 Published 28 May 2020
Abstract
DYRK1A (dual specificity tyrosine-(Y)-phosphorylation-regulated kinase 1 A) is a high
confidence autism risk gene that encodes a conserved kinase. In addition to autism, patients with putative loss of function variants in DYRK1A exhibit microcephaly, intellectual disability, developmental delay, and/or congenital anomalies of the kidney and urinary tract. DYRK1A is also located within the critical region for Down syndrome; therefore, understanding the role of DYRK1A in brain development is crucial for understanding the pathobiology of multiple developmental disorders. To characterize the function of this gene, we used the diploid frog, Xenopus tropicalis. We discover that Dyrk1a is expressed in ciliated tissues, localizes to ciliary axonemes and basal bodies, and is required for ciliogenesis. We also demonstrate that Dyrk1a localizes to mitotic spindles and that its inhibition leads to decreased forebrain size, abnormal cell cycle progression, and cell death during brain development. These findings provide hypotheses about potential mechanisms of pathobiology and underscore the utility of X. tropicalis as a model system for understanding neurodevelopmental disorders.
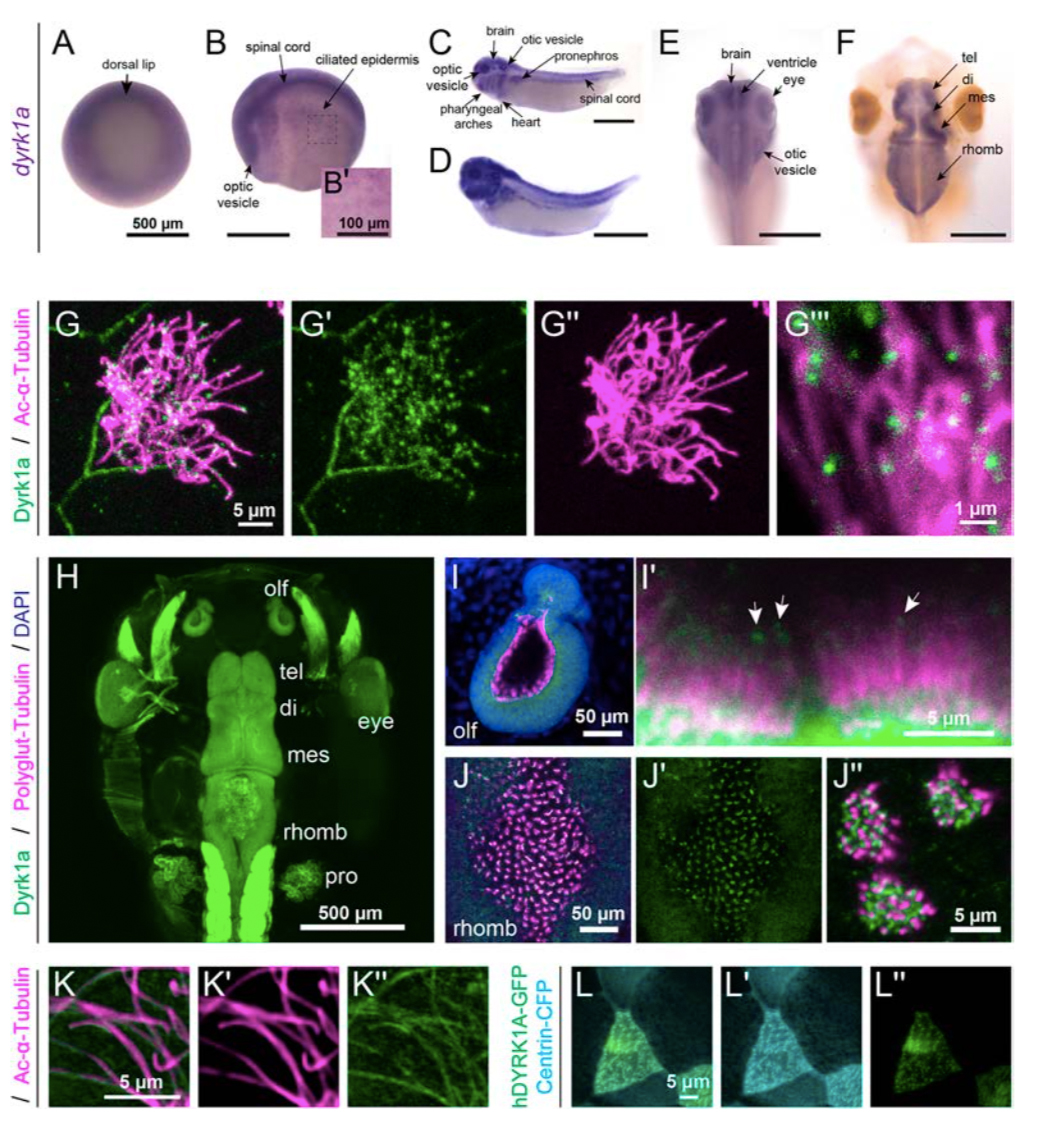
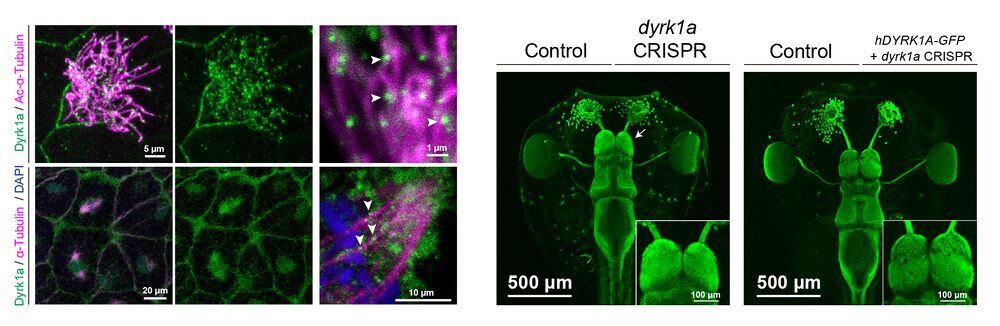
Figure 1. Expression and localization of Dyrk1a during X. tropicalis development.
(A-F): By RNA in situ hybridization in X. tropicalis, dyrk1a is expressed during gastrulation (stage 10.5, A) and at stage 20 (B) in the brain, spinal cord, optic vesicles, and ciliated epidermis (B’). At stages 32 (C) and 35 (D), dyrk1a is expressed in the brain, spinal cord, otic vesicles, optic vesicles, pharyngeal arches, heart, epidermis, and pronephros. At stages 40 (E) and 46 (F), dyrk1a brain expression, especially along ventricles. T elencephalon (tel), diencephalon (di), mesencephalon (mes), and rhombencephalon (rhomb). Scale is 500 μm, except 100 μm in B’. (G) Embryonic epidermis antibody staining for Dyrk1a protein (green) shows puncta along ciliary axonemes labeled by acetylated α-Tubulin (Ac-α-Tubulin, magenta). See Fig. S1 for antibody validation. (H) Dorsal view of whole-mount antibody staining showing Dyrk1a (green) throughout the nervous system and in the pronephros (pro). (I-J) Dyrk1a (green) is on cilia marked by polyglutamylated-Tubulin (polyglut-Tubulin, magenta) in the olfactory epithelium (olf, I) and in the rhombencephalon (J). White arrows (I’) indicate puncta of Dyrk1a (green) on cilia. Also note strong membrane staining. (K) Human GFP-tagged DYRK1A (hDYRK1A-GFP, green) localizes in puncta along ciliary axonemes labeled by acetylated α- Tubulin (magenta). (L) hDYRK1A-GFP (green) co-localizes with Centrin-CFP (cyan) at basal bodies.

Figure 2. dyrk1a is required for ciliogenesis and brain size.
(A-C) Stage 35 X. tropicalis embryonic epidermis stained for acetylated α-Tubulin (cilia, magenta), phalloidin (actin, green), and expressing Centrin-CFP (basal bodies, blue). Injection of dyrk1a morpholino (MO) (B) or CRISPR/Cas9 reagents (C) causes a loss in cilia (magenta) compared to the injected control (A). See also Fig. S2-S3. (D) Quantification of ciliogenesis phenotype (severe, mild, and normal) in multiciliated cells (MCCs) by condition. n > 35 for each condition. (E-H) Dorsal view of β-Tubulin antibody staining (green) of X. tropicalis tadpoles, injected (right side) with dyrk1a morpholino (MO) (E), dyrk1a CRISPR (F), or co-injected with dyrk1a CRISPR and human GFP-tagged DYRK1A (hDYRK1A-GFP) (H). Dyrk1a kinase inhibitor harmine-treated embryos are affected bilaterally (1.25 μM, G). Telencephalon region shown as insets. White arrows point to telencephalon size phenotypes. (I) Quantification of log2 fold change of telencephalon size normalized to control. Box plot whiskers are maximum and minimums, boxes are interquartile range, and line is median. Every point is an animal. p-values are from non-parametric Mann-Whitney rank sum tests compared to negative control CRISPR targeting pigmentation gene slc45a2. For harmine, comparison to paired DMSO treatment also gives a p-value < 0.0001. ** indicates p < 0.01; **** indicates p < 0.0001; “n.s.” indicates “not significant”, p > 0.05.
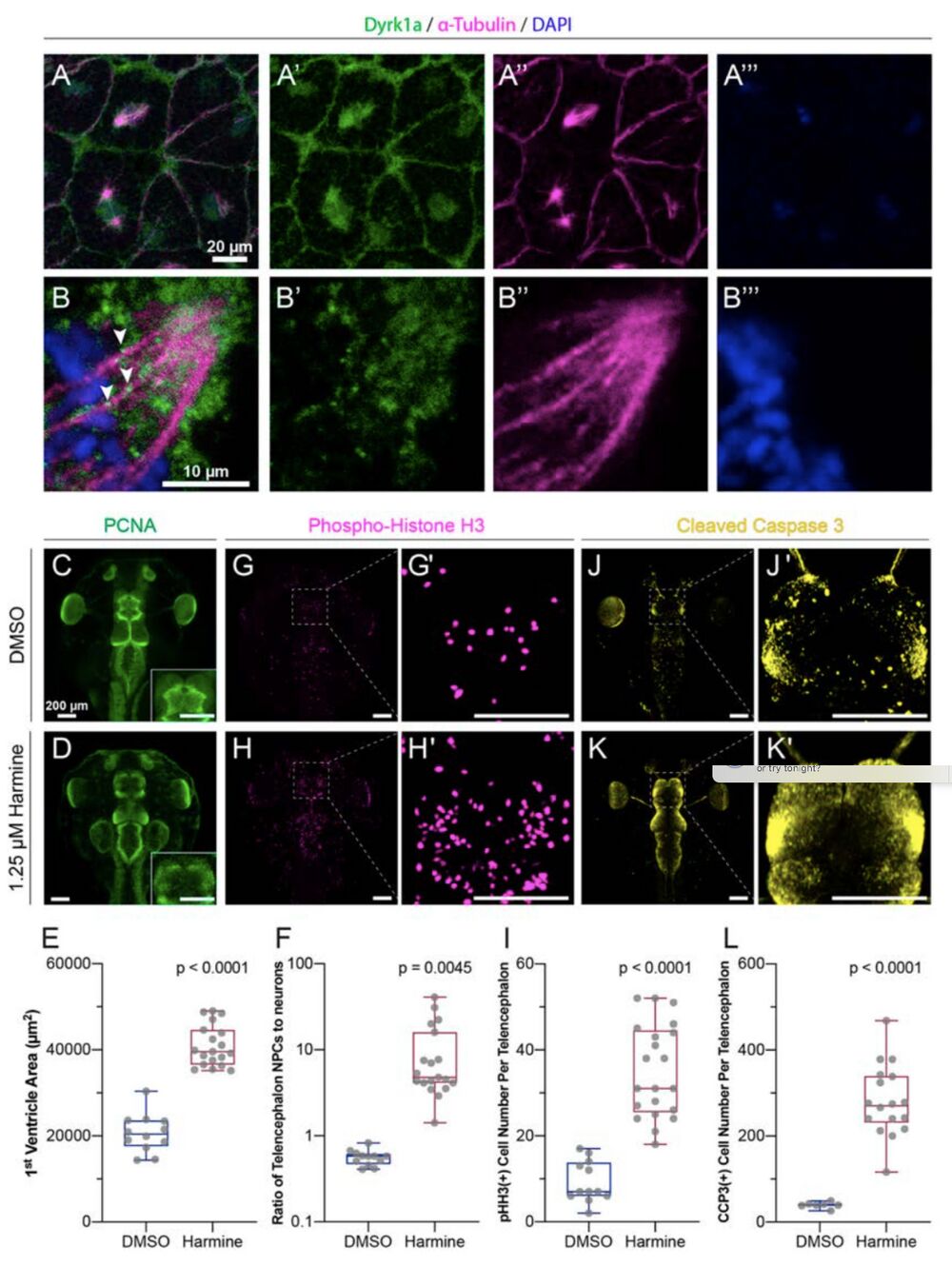
Figure 4: Dyrk1a localizes to mitotic spindles and is required for cell cycle progression and survival.
(A-B) Stage 10 embryos stained for Dyrk1a (green), α-Tubulin (microtubules, magenta), and DAPI (DNA, blue). White arrows indicate Dyrk1a puncta near mitotic spindle. (C- L) Dorsal view of X. tropicalis tadpoles treated with DMSO (top row) or 1.25 μM Dyrk1a inhibitor Harmine (bottom row). (C-D) Antibody staining for PCNA (proliferating cell nuclear antigen, S phase marker, green). Telencephalon region shown as an inset. (E) Quantification of first ventricle area from PCNA staining. (F) Quantification of the ratio of neural progenitor cells (NPCs, PCNA area) to the area of differentiated neurons in each telencephalon (log scale). (G- H) Antibody staining for pHH3 (phospho-histone H3, M phase marker, magenta). (G’, H’) High magnification views of telencephalon regions from G and H. (I) Quantification of pHH3 positive cell number per telencephalon. See also Fig. S4. (J-K) Antibody staining for CCP3 (cleaved caspase 3, cell death marker, yellow). (J’, K’) High magnification view of telencephalon from J and K. (L) Quantification of CCP3 positive cell number per telencephalon. Scale bars in C-L including insets are 200 μm. Box plot whiskers are maximum and minimum, boxes are interquartile range, and line is median. Every point is an animal. p-values are from non- parametric Mann-Whitney rank sum tests.
Adapted with permission from Development on behalf of The Company of Biologists: Willsey & Xu et al. (2020). Neurodevelopmental disorder risk gene DYRK1A is required for ciliogenesis and brain size in Xenopus embryos. Development 2020 : dev.189290 doi: 10.1242/dev.189290 Published 28 May 2020.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2020-06-01