Long-term transcription factor binding stabilizes gene expression
Long-term association of a transcription factor with its chromatin binding site can stabilize gene expression and cell fate commitment
J. B. Gurdon, Khayam Javeda, Munender Vodnalab, and Nigel Garretta
Click here to view article at PNAS.
Click here to view article on Pubmed.
Click here to view article on Xenbase.
Abstract
Some lineage-determining transcription factors are overwhelmingly important in directing embryonic cells to a particular differentiation pathway, such as Ascl1 for nerve. They also have an exceptionally strong ability to force cells to change from an unrelated pathway to one preferred by their action. Transcription factors are believed to have a very short residence time of only a few seconds on their specific DNA or chromatin-binding sites. We have developed a procedure in which DNA containing one copy of the binding site for the neural-inducing factor Ascl1 is injected directly into a Xenopus oocyte nucleus which has been preloaded with a limiting amount of the Ascl1 transcription factor protein. This is followed by a further injection of DNA as a competitor, either in a plasmid or in chromosomal DNA, containing the same binding site but with a different reporter. Importantly, expression of the reporter provides a measure of the function of the transcription factor in addition to its residence time. The same long residence time and resistance to competition are seen with the estrogen receptor and its DNA response elements. We find that in this nondividing oocyte, the nerve-inducing factor Ascl1 can remain bound to a specific chromatin site for hours or days and thereby help to stabilize gene expression. This stability of transcription factor binding to chromatin is a necessary part of its action because removal of this factor causes discontinuation of its effect on gene expression. Stable transcription factor binding may be a characteristic of nondividing cells.
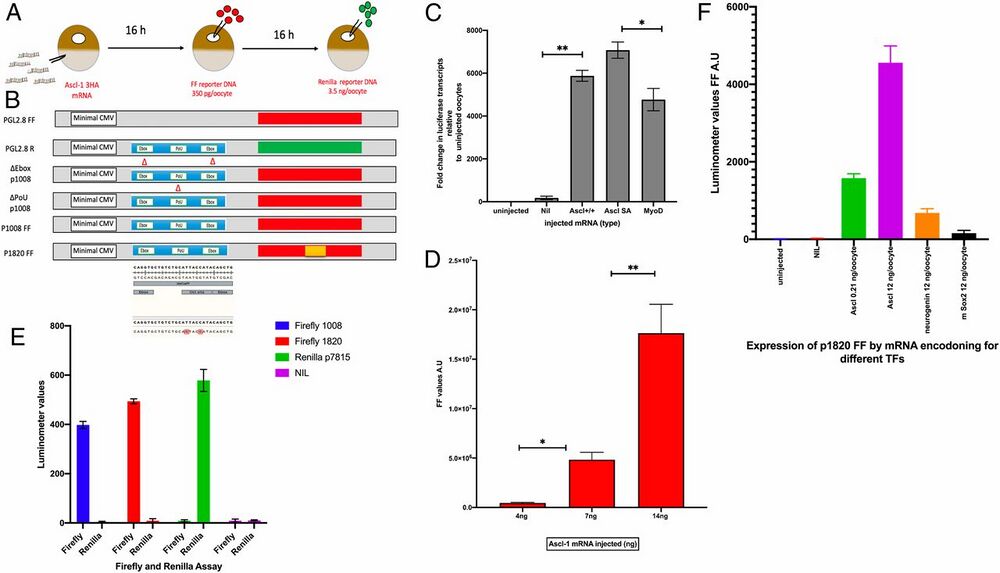
Fig. 1.
Experimental design. (A) The sequence of injections on living oocytes of Xenopus. The normal injection volume is 14 nL, aimed for the cytoplasm (mRNA) or the nucleus (GV). Nucleic acids were made up in water for injection. (B) DNA plasmid composition. The various plasmids used are derived from PGL4.28 (Promega). P1008 was provided by A. Philpott (Jeffrey Cheah Biomedical Centre Cambridge Biomedical Campus, Cambridge, UK) (ref. 25). The base pairs marked in red had been mutated as shown for the E-box and Pou sequences. (C) The mRNAs shown cause a substantial increase in the abundance of transcripts from the E-box–Pou–E-box domain of p1008 DNA. Ascl1 SA is a variant of mouse Ascl1 in which alanines have been mutated to serines. *P = 0.001; **P = 0.009. (D) Different amounts and kinds of Ascl1 mRNA induce a huge increase in Firefly expression of p1008 DNA. *P = 0.005; **P = 0.007. (E) The reporters FF for p1008 and p1820 respond strongly and independently to Ascl1 mRNA. The reporter Renilla responds similarly to DNA p7815 R. (F) Different kinds of mRNA induce specific reporter responses. Neurogenin (orange) has almost no effect and Sox2 (black) of mice and no effect at all on Firefly expression (red). Error bars represent the SEM in three independent experiments (n = 3).
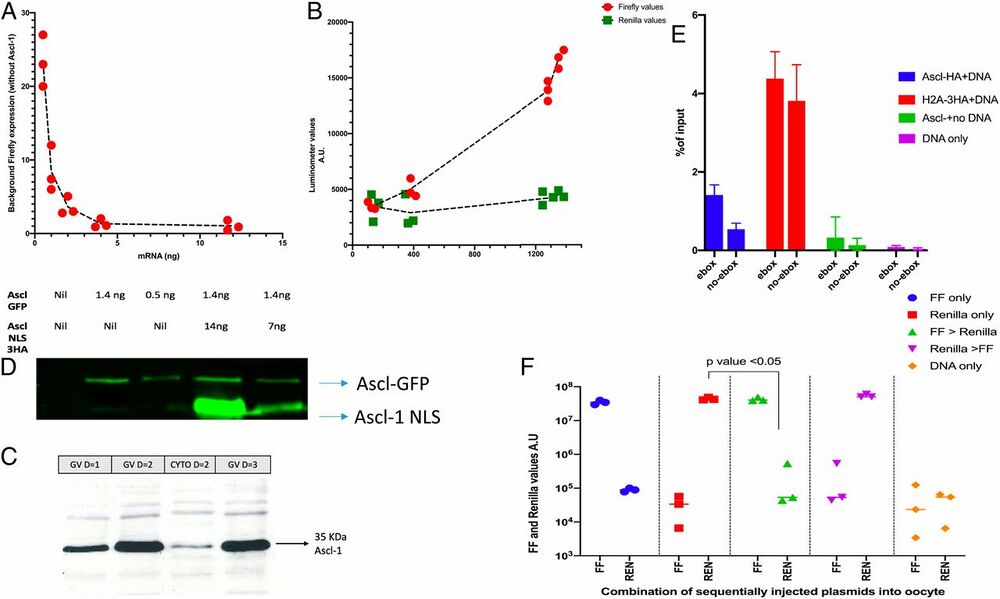
Fig. 2.
Characterization of gene expression induced by Ascl1 mRNA in DNA-injected oocytes. (A) The background level of luciferase in oocytes that received DNA but no mRNA was 1 to 10% of the mRNA-induced level. (B) High levels of injected Ascl1 mRNA are sufficient to induce expression of a second injected DNA. (C) Ascl1 mRNA-encoded protein concentrates in the GV of injected oocytes and remains at a high level for 3 d. (D) A nuclear localization signal in Ascl1 mRNA helps to increase GV localization injected oocytes. (E) Injected DNA soon becomes associated with histone from injected histone mRNA. (F) The order of addition of DNA (FF or R first or sequential) gives the same conclusion: The first DNA dominates over the second. Error bars shows SEM for (n = 3) where applicable.
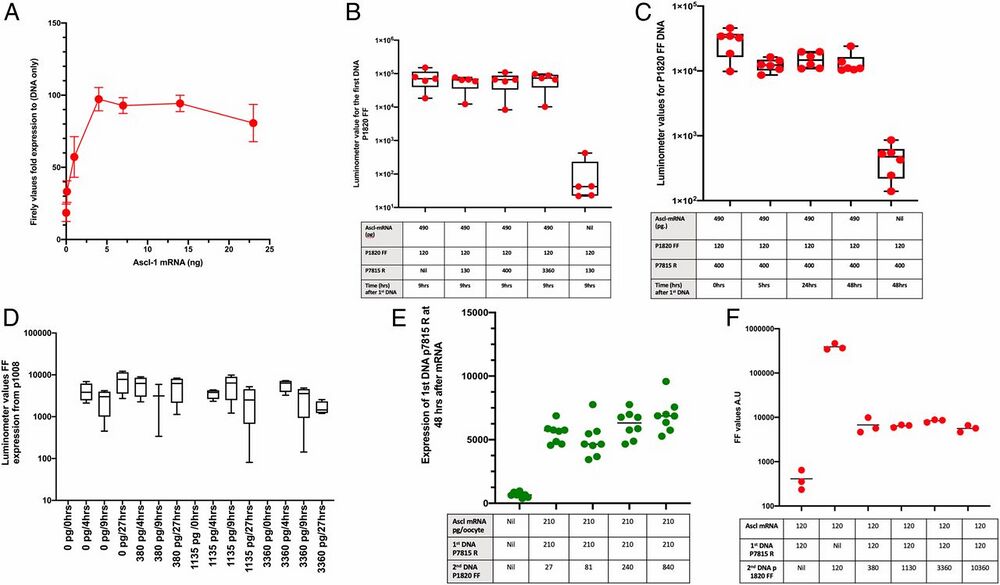
Fig. 3.
Lack of competition for sequentially introduced DNA binding sites. (A) Fifty picograms of plasmid DNA are nearly saturated by 4 ng of Ascl1 mRNA. In most samples of oocytes, 500 pg to 1 ng of Ascl1 mRNA per oocyte containing 210 pg of p1820 DNA was sufficient to ensure that the synthesized protein limits the amount of luciferase reporter seen. Beyond this level, no further reporter expression is obtained by injecting a further amount of DNA or RNA. (B) Increasing the amount of a second competitor DNA up to 3,360 pg/oocyte does not increase the amount of the first DNA’s reporter (p1820 FF). (C) A lack of competition is seen by a second DNA if the duration of incubation is prolonged for 2 d after the first injection. (D) Summary of results from 17 frogs in different experiments, showing that no significant reporter expression is obtained from a second injection of DNA. The synthesized DNA was injected at 0.2 to 12 ng/oocyte. (E) After Ascl1 mRNA at 210 pg/oocyte, we inject competitor DNA 7815R, also at 210 pg/oocyte. The next day, expression p1820 FF is injected at concentrations from nil to 840 pg/oocyte. (F) The amount of expression DNA p 1820 FF is the same in all samples, but the amount of p7815R as first DNA, as a competitor, shows the expected increase.
Adapted with permission from PNAS on behalf of the National Academy of Sciences: Gurdon et al. (2020). Long-term association of a transcription factor with its chromatin binding site can stabilize gene expression and cell fate commitment. PNAS first published June 12, 2020 https://doi.org/10.1073/pnas.2000467117.
Last Updated: 2020-06-23