New Xenopus Genome Assemblies
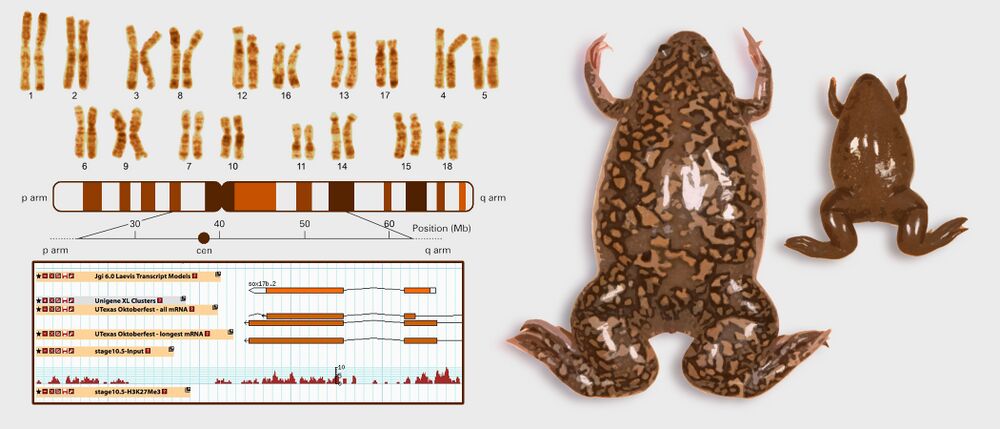
The International Xenopus Genome Consortium (including groups from the US, Japan, France and the UK) have made the newest Xenopus laevis v7.1 and Xenopus tropicalis v8.0 genome assemblies, from the Rokhsar group, available to the public through Xenbase.
Xenopus laevis v7.1 has improved contiguity with 2.7 Gb total scaffold length and 50% of the assembly in contigs >19.3kb.
Xenopus tropicalis v8.0 was generated using improved computational methods, incorporating additional BAC-end data and by integrating a dense genetic map resulting in a more contiguous assembly mapped to chromosomes with 50% of the contigs >72 kb in length.
We are in the process of integrating the new assemblies into Xenbase.
- Currently laevis v7.1 and tropicalis v8.0 are available for FTP download
- X. laevis v7.1 can also be accessed by BLAST+ and on the Gbrowse.
- The best way to query laevis v7.1 at the moment is via BLAST+
Image of X. laevis chromosome karyotype adapted with permission from Macmillan Publishers Ltd: Uno et al. (2013). Homoeologous chromosomes of Xenopus laevis are highly conserved after whole-genome duplication. Heredity: copyright (2013)
Image of frogs adapted from the Amaya lab.
Last Updated: 2013-12-11