RNA whole-mount in situ hybridization proximity ligation assay (rISH-PLA)
RNA Whole-Mount In situ Hybridisation Proximity Ligation Assay (rISH-PLA), an Assay for Detecting RNA-Protein Complexes in Intact Cells
Ioannis M. Roussis, Matthew Guille, Fiona A. Myers*, Garry P. Scarlett*
PLOS One (2016) - link to Research Article
Link to protocol on Xenbase Protocols Wiki
Download Word document of full rISH-PLA protocol
Abstract
Techniques for studying RNA-protein interactions have lagged behind those for DNA-pro- tein complexes as a consequence of the complexities associated with working with RNA. Here we present a method for the modification of the existing In Situ Hybridisation–Proxim- ity Ligation Assay (ISH-PLA) protocol to adapt it to the study of RNA regulation (rISH-PLA). As proof of principle we used the well-characterised interaction of the Xenopus laevis Stau- fen RNA binding protein with Vg1 mRNA, the complex of which co-localises to the vegetal pole of Xenopus oocytes. The applicability of both the Stau1 antibody and the Locked Nucleic Acid probe (LNA) recognising Vg1 mRNA were independently validated by whole- mount Immunohistochemistry and whole-mount in situ hybridisation assays respectively prior to combining them in the rISH-PLA assay. The rISH-PLA assay allows the identifica- tion of a given RNA-protein complex at subcellular and single cell resolution, thus avoiding the lack of spatial resolution and sensitivity associated with assaying heterogenous cell populations from which conventional RNA-protein interaction detection techniques suffer. This technique will be particularly usefully for studying the activity of RNA binding proteins (RBPs) in complex mixtures of cells, for example tissue sections or whole embryos.
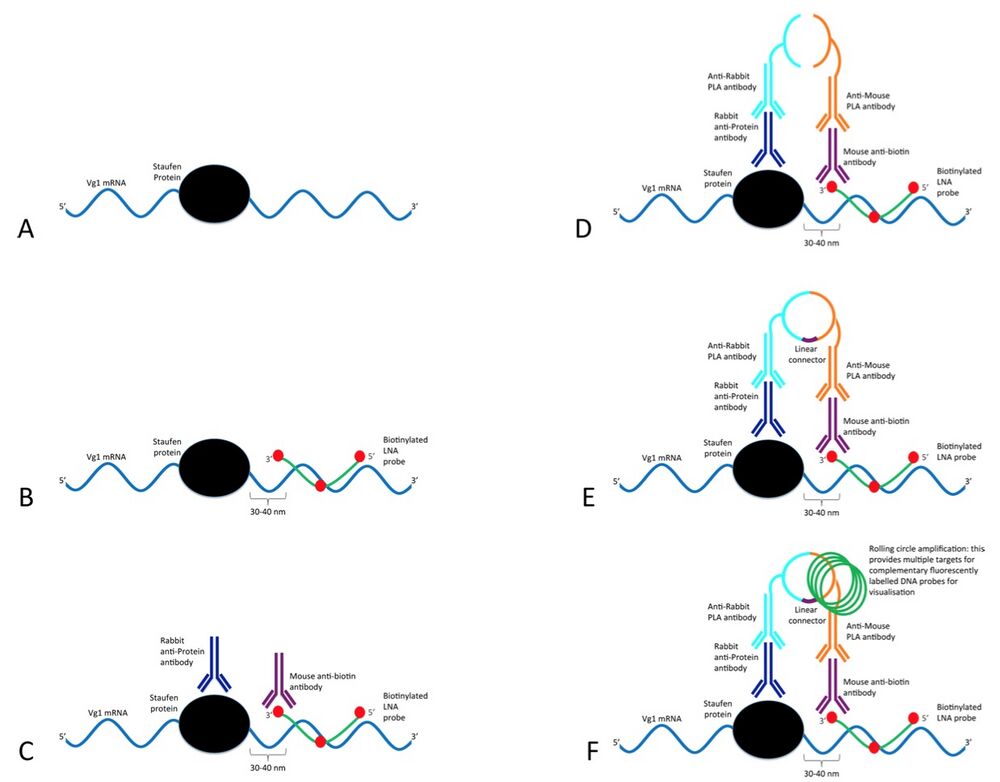
Schematic of the ISH-PLA modified for use with RNA (rISH-PLA).
The ISH-PLA technique has been developed for the analysis of the co- localisation of protein complexes and specific DNA sequences. In this paper the method is adapted for RNA. (A) In vivo interaction of Vg1 mRNA (blue) with Xenopus Staufen (black). (B) In the first step a biotinylated LNA probe (green with biotin indicated in red) targets a specific RNA sequence close to the binding site of XStau1 in the Vg1 mRNA is added (see results section for details on LNA probe design). (C) Two primary antibodies raised in different species are added. In this case a rabbit antibody (dark blue) targets the XStau1 and a mouse antibody (purple) targets the biotin label in the LNA probe. (D) Two secondary PLA species-specific antibodies (light blue and orange) conjugated to oligonucleotides are then added. Since the LNA probe and the protein are in close proximity the secondary antibodies and their conjugated oligonucleotides are consequently also in close proximity. (E) The conjugated oligonucleotides are joined and circularised by DNA ligation upon introduction of linear connector oligonucleotide (dark purple). (F) The two oligonucleotides then commence rolling circle replication (the amplified circular DNA molecule is annotated in green). After the amplification reaction, fluorescent labelled complementary oligonucleotide probes are added to highlight the product.
Last Updated: 2016-02-10