The Epigenetic Control of Pluripotent Signal Competence
Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals
Nature Communications Volume 10, Article number: 4269 (2019)
George E. Gentsch, Thomas Spruce, Nick D. L. Owens & James C. Smith
Gentsch et al. systematically investigated the role of sequence-specific transcription factors in establishing signal competence, a long-standing biological property (Conrad Waddington, 1940) that determines the way a cell responds to inductive signals. They show that maternal pluripotency factors initiate the epigenetic remodelling of thousands of gene regulatory elements upon which signal mediators can act to regionalise zygotic genome activity and specify the three germ layers. Their discovery is a first step in understanding how proteins permit a cell type-specific signal response, which will increase the success of engineering patient-specific tissue for the restoration of damaged organs.
Click here to view article at Nature Communications.
Abstract
Embryonic development yields many different cell types in response to just a few families of inductive signals. The property of signal-receiving cells that determines how they respond to inductive signals is known as competence, and it differs in different cell types. Here, we explore the ways in which maternal factors modify chromatin to specify initial competence in the frog Xenopus tropicalis. We identify early-engaged regulatory DNA sequences, and infer from them critical activators of the zygotic genome. Of these, we show that the pioneering activity of the maternal pluripotency factors Pou5f3 and Sox3 determines competence for germ layer formation by extensively remodelling compacted chromatin before the onset of inductive signalling. This remodelling includes the opening and marking of thousands of regulatory elements, extensive chromatin looping, and the co-recruitment of signal-mediating transcription factors. Our work identifies significant developmental principles that inform our understanding of how pluripotent stem cells interpret inductive signals.
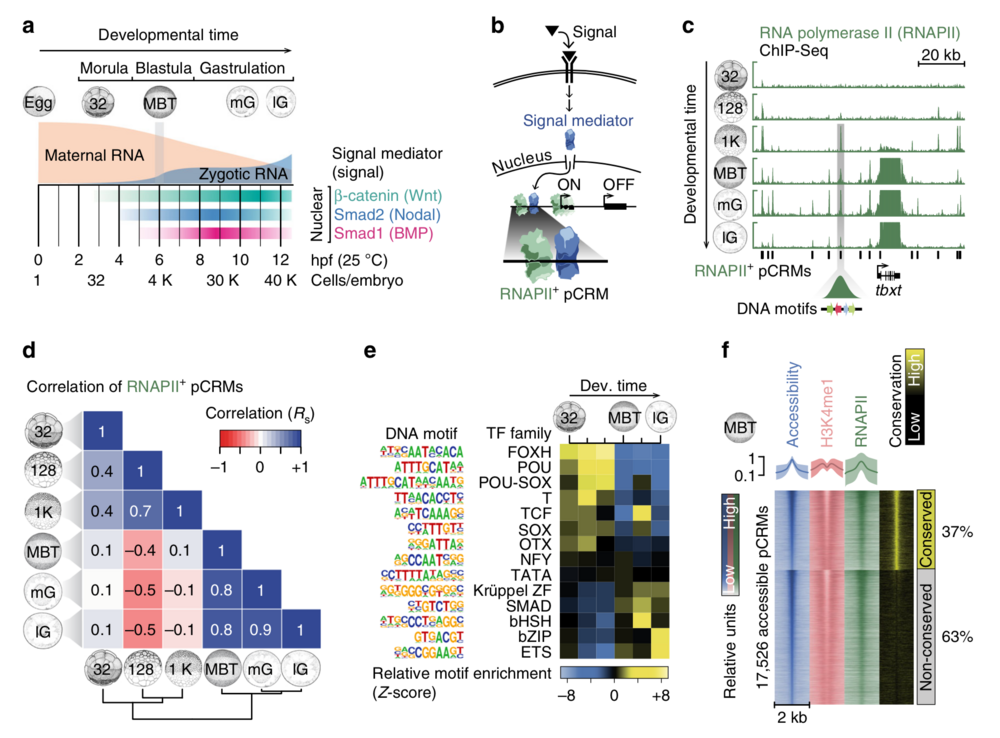
Fig. 1 Characterisation of pCRMs instructing ZGA. a Timeline (hpf, hours post-fertilisation, at 25 °C) of the maternal-to-zygotic transition and earliest signalling events (nuclear accumulation of Wnt, Nodal and Bmp signal mediators β-catenin, Smad2 and Smad1, respectively) during early X. tropicalis development up to the late gastrula embryo (12 hpf) with ∼40,000 (40 K) cells. b Signal transduction pathway causing signal mediators to enter the nucleus and engage with pCRMs (e.g., marked by RNAPII). c Snapshot of RNAPII recruitment to pCRMs of the zygotic gene locus tbxt from the 32-cell to the late gastrula stage. The underlying DNA sequence of RNAPII+ pCRMs are used to discover enriched DNA motifs de novo (illustrated as coloured arrows for one RNAPII+ pCRM). d Spearman correlations (Rs) of RNAPII binding levels across ∼27,000 pCRMs (Supplementary Data 2) between the indicated developmental stages. e Temporal enrichment (Z-score) of consensus DNA motifs known to be recognised by indicated TF families among RNAPII+ pCRMs. f MBT-staged heat map of DNase-probed chromatin accessibility, RNAPII binding and H3K4me1 marking (n = 2 biologically independent samples) across ∼17,500 pCRMs (Supplementary Data 3) grouped by sequence conservation levels (phastCons) and sorted by the statistical significance of pCRM accessibility. Abbreviations used for the developmental timeline: 32, 128 and 1 K, 32-, 128- and 1024-cell stage; MBT, mid-blastula transition; eG, mG and lG, early, mid- and late gastrula stage
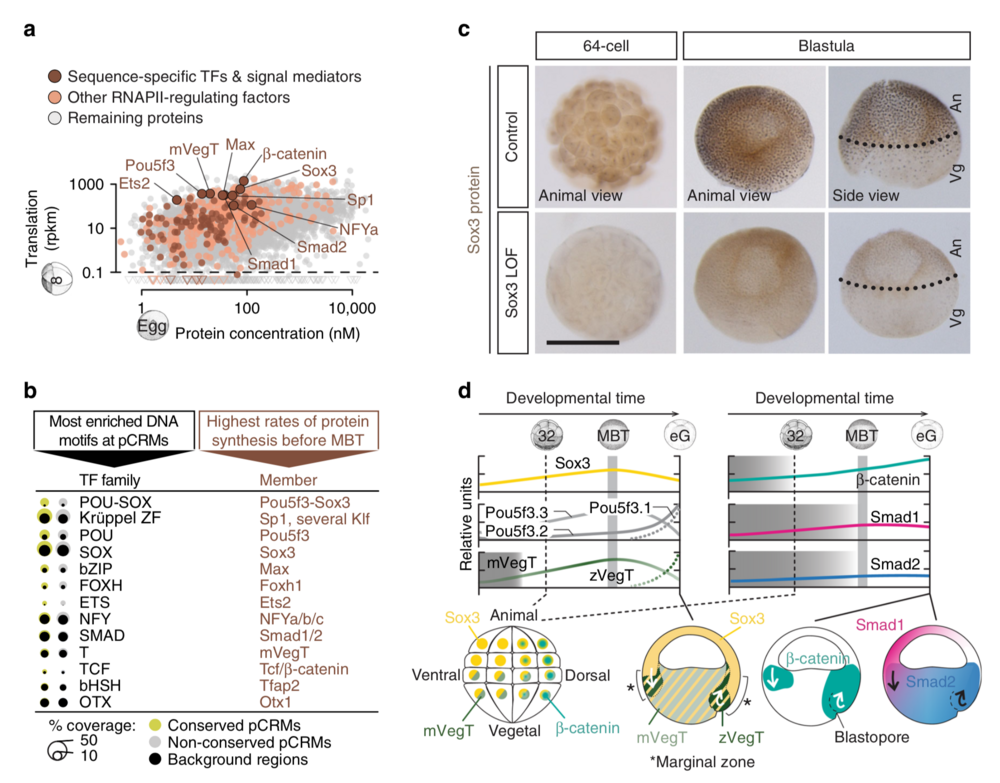
Fig. 2 Search for ZGA-critical proteins based on their significantly enriched DNA recognition motifs at accessible and engaged (RNAPII+/H3K4me1+) pCRMs and their high translation frequency before the MBT. a Maternal protein concentrations in the egg13 versus ribosome footprint (translation) levels at the 8-cell stage14. Most frequently translated representatives of various TF families are labelled. b Matching canonical pCRM-enriched DNA motifs (sorted by statistical significance) with frequently translated TFs and signal mediators. c WMIHC of Sox3 protein in control and Sox3 loss-of-function (LOF) embryos at the 64-cell and blastula stage. Nuclear accumulation of Sox3 protein was detected in both the animal (An) and vegetal (Vg) hemisphere of control embryos. Scale bar, 0.5 mm. d Graphical illustration of protein levels (derived from mass spectrometry data42) and nuclear localisations (mainly derived from WMIHC, see references below) of selected TFs and signal mediators based on our and previously published results: Sox3 (this study and ref. 50), mPouV (Pou5f3.2 and Pou5f3.3) and (zygotic) Pou5f3.1 (deduced from transcript data15), mVegT and zVegT16, β-catenin10,17, Smad1 (this study and refs. 10,18) and Smad2 (this study and refs. 10,18). Shaded boxes indicate periods of non-nuclear protein localisation. Arrows indicate tissue movements of gastrulation. Abbreviations used for the developmental timeline: 8 and 32, 8-cell and 32-cell stage; MBT, mid-blastula transition; and eG, early gastrula stage
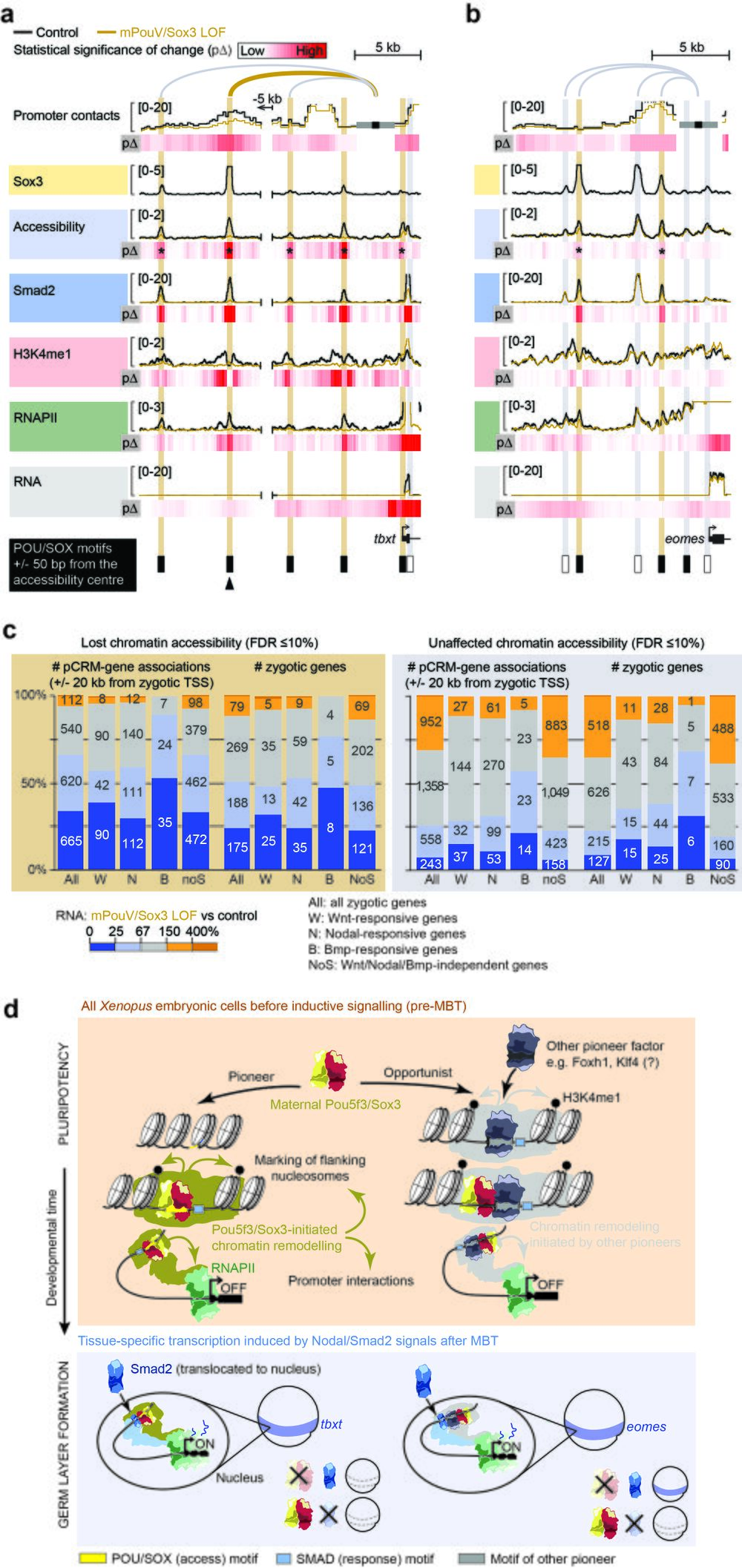
Fig. 10 Maternal Pou5f3/Sox3-initiated chromatin remodelling to prime the first transcriptional response to inductive signals during ZGA. a, b Superimposed line tracks show promoter contact frequencies, chromatin accessibilities and DNA occupancies of various chromatin components (Smad2, H3K4me1 and RNAPII) at the Nodal-responsive mesoderm determinants tbxt (a) and eomes (b) between control (uninjected) and mPouV/Sox3 LOF embryos. Heat maps (pΔ) below each superimposed line track show the statistical significance (Wald test) of changes caused by mPouV/Sox3 LOF. The footer highlights the occurrences of canonical POU/SOX motifs (black filled rectangles) at accessible pCRMs (±50 bp from the accessibility centre) and one strongly affected pCRM with an arrowhead. Asterisks on the pΔ heat map mark significant (FDR ≤10%) reductions to pCRM accessibility. pCRMs are boxed in and their frequency of contacts with the promoter are illustrated with an arc of varying strength. Boxes of affected pCRM and arcs of promoter contacts are coloured orange. c Stacked bar graphs summarise the correlation of unaffected and significantly reduced (FDR ≤10%) pCRM accessibility with RNAPII-mediated expression of all, signal responsive and non-responsive zygotic genes in mPouV/Sox3 LOF embryos at the MBT. These correlations and corresponding numbers (placed on the stacked bars) are shown for pCRM-gene associations and zygotic genes. TSS-centric maps of reduced chromatin accessibility are shown for signal responsive and non-responsive genes in Supplementary Figs. 15–17. d Model of chromatin pioneering and opportunistic engagement to predefine first zygotic responses to inductive signals
Adapted with permission from Springer Nature on behalf of Cell Nature Communications: Gentsch et al. (2019). Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals
Nature Communications Volume 10, Article number: 4269 (2019) Copyright (2019).
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2019-09-19