Uncovering selective cooperation between the retinoblastoma and p53 cancer pathways in X. tropicalis
RBL1 (p107) functions as tumor suppressor in glioblastoma and small-cell pancreatic neuroendocrine carcinoma in Xenopus tropicalis.
Thomas Naert, Dionysia Dimitrakopoulou, Dieter Tulkens, Suzan Demuynck, Marjolein Carron, Rivka Noelanders, Liza Eeckhout, Gert Van Isterdael, Dieter Deforce, Christian Vanhove, Jo Van Dorpe, David Creytens & Kris Vleminckx
Oncogene (2020). https://doi.org/10.1038/s41388-020-1173-z
Click here to view article at Oncogene.
Click here to view article on Pubmed.
Click here to view article on Xenbase.
The study first describes stable tp53 heterozygote and homozygote knockout lines in Xenopus tropicalis that develop leukemia and malignant sarcoma, thereby establishing the first frog model for the Li-Fraumeni Syndrome. Subsequently, mosaic mutant animals (crispants) were obtained by CRISPR/Cas9 multiplexed genome editing to investigate cooperation of the retinoblastoma and the p53 cancer pathways during cancer formation. This revealed the induction of several cancer types, including choroid plexus carcinoma, pancreatic neuroendocrine carcinoma and glioma, that developed with high penetrance and short latency. Using targeted amplicon sequencing, the mutation status of the rb1, rbl1 and tp53 genes was investigated in the tumors, thereby uncovering the mutational events required for each tumor type. Finally, quadruple CRISPR gene editing (rb1/rbl1/tp53/pten) in the neural lineage induced very penetrant and highly aggressive glioblastoma with a latency of less than 35 days. These data further encourage the use of Xenopus tropicalis as a valuable cancer model that can be used to identify novel cancer driver genes and uncover potential genetic dependencies.
Abstract
Alterations of the retinoblastoma and/or the p53 signaling network are associated with specific cancers such as high-grade astrocytoma/glioblastoma, small-cell lung cancer (SCLC), choroid plexus tumors, and small-cell pancreatic neuroendocrine carcinoma (SC-PaNEC). However, the intricate functional redundancy between RB1 and the related pocket proteins RBL1/p107 and RBL2/p130 in suppressing tumorigenesis remains poorly understood. Here we performed lineage-restricted parallel inactivation of rb1 and rbl1 by multiplex CRISPR/Cas9 genome editing in the true diploid Xenopus tropicalis to gain insight into this in vivo redundancy. We show that while rb1 inactivation is sufficient to induce choroid plexus papilloma, combined rb1 and rbl1 inactivation is required and sufficient to drive SC-PaNEC, retinoblastoma and astrocytoma. Further, using a novel Li-Fraumeni syndrome-mimicking tp53 mutant X. tropicalis line, we demonstrate increased malignancy of rb1/rbl1-mutant glioma towards glioblastoma upon concomitant inactivation of tp53. Interestingly, although clinical SC-PaNEC samples are characterized by abnormal p53 expression or localization, in the current experimental models, the tp53 status had little effect on the establishment and growth of SC-PaNEC, but may rather be essential for maintaining chromosomal stability. SCLC was only rarely observed in our experimental setup, indicating requirement of additional or alternative oncogenic insults. In conclusion, we used CRISPR/Cas9 to delineate the tumor suppressor properties of Rbl1, generating new insights in the functional redundancy within the retinoblastoma protein family in suppressing neuroendocrine pancreatic cancer and glioma/glioblastoma.
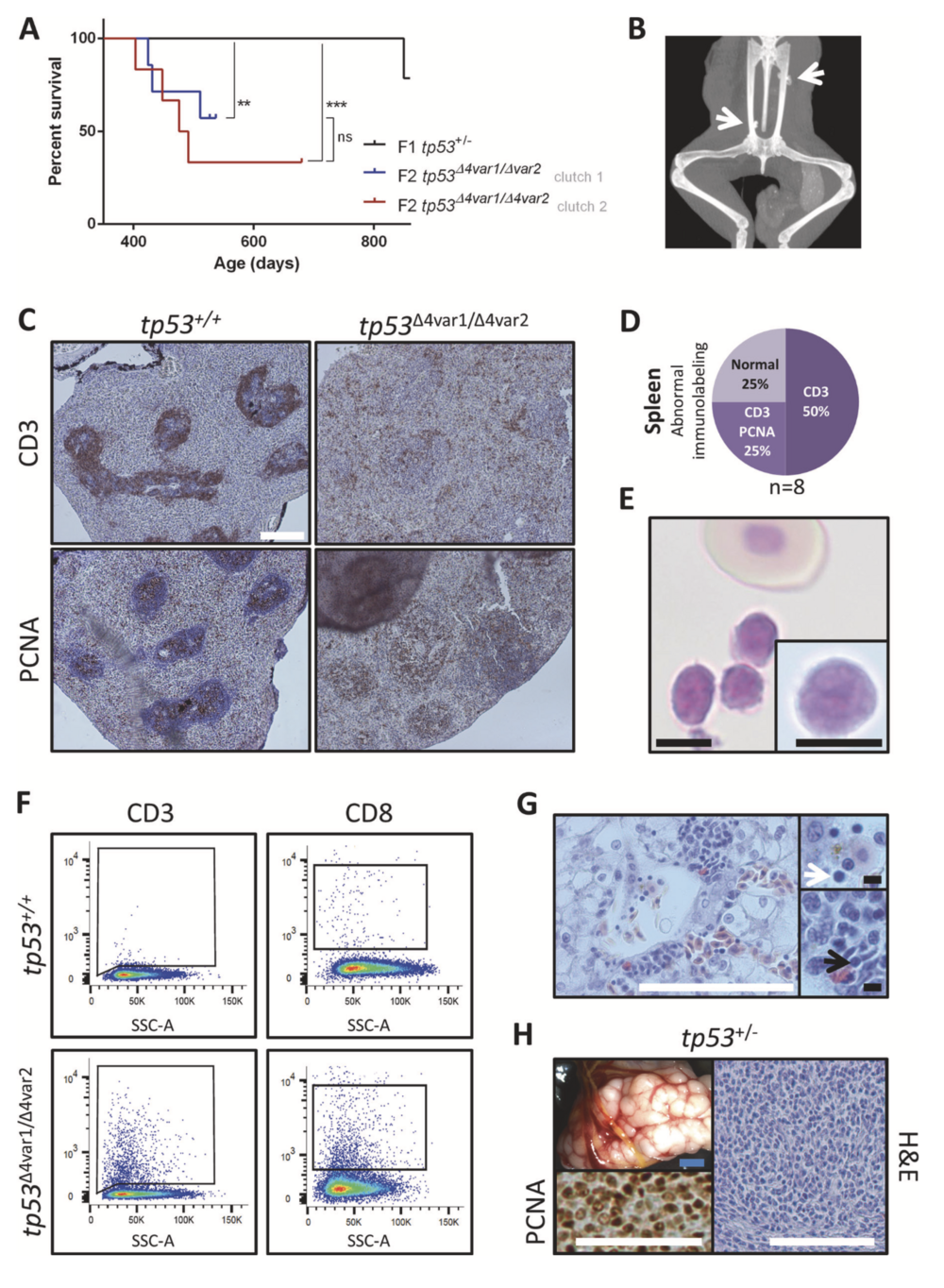
Fig. 1 Tp53 mutant X. tropicalis develop hematological malignancy and sarcomas. a Kaplan–Meier survival analysis on cohorts consist- ing of two clutches of tp53 homozygous knockout (n = 6; red and n = 7; blue) and one clutch of heterozygous tp53 knockout (n = 14; black) X. tropicalis. Statistical analysis was done using the Prism Mantel–Cox test (ns not significant; **p < 0.01; ***p < 0.001). Chi- Squared values and the Hazard ratios are listed in Supplementary table 2A. b X-ray imaging of a 28-month old tp53+/Δ4var2 demonstrating ectopic calcified structures (white arrows). c (Left panels) In the wild- type spleen, CD3+ T-cells form a ring-like structure around the PCNA+ B-cells located in the white pulp. (Right panels) Disruption of a normal CD3+ ring-like structure observed in a tp53Δ4var2/Δ4var2 animal is indi- cative of hematological malignancy. d Pie chart summarizing the observed splenic immunostaining for CD3 and PCNA in tp53Δ4var1/Δ4var2 animals. Staining patterns being either normal or with loss of the typical CD3+ ring structure (CD3) with or without ectopic staining of PCNA (CD3/PCNA) in the red pulp. e Photomicrograph of Natt–Herrick stained blood demonstrating a cluster of leukocytes and a normal nucleated erythrocyte. Inset demonstrates a high-magnification photo- micrograph of a lymphoblast. f Flow cytometry analysis reveals an increased number of both CD3+ (left) and CD8+ lymphoblasts (right) in peripheral blood of a tp53Δ4var1/Δ4var2 animal, when compared with an age-matched control. g Histopathology of the liver reveals diffuse infiltration of T-lymphoblasts, also enriched within the liver capillary (white arrow; top inset) and in between the liver parenchymal cells (black arrow; bottom inset). h Sarcoma in a tp53+/Δ4var2 animal observed upon gross examination, with associated histopathology and demon- stration of malignant nature by PCNA proliferation staining. White scale bar is 100 μm and black scale bar is 5 μM.

Fig. 2 Small-cell pancreatic neuroendocrine carcinoma (SC- PaNEC) in X. tropicalis upon mosaic CRISPR/Cas9 genome edit- ing of rb1 and rbl1 in the anterior endoderm. a Breeding and CRISPR/Cas9 injection scheme demonstrating the generation of the single mutant KO (smKO—orange) (rb1 CRISPR/Cas9 injection in embryos obtained from a tp53Δ4var1/+ × tp53Δ4var2/+ intercross), double mutant KO (dmKO—blue) (rb1/rbl1 CRISPR/Cas9 injection in embryos obtained from a tp53Δ4var1/+ × tp53Δ4var2/+ intercross) and triple mutant KO (tmKO—purple) (rb1/rbl1/tp53 CRISPR/Cas9 injection in embryos obtained from intercrossing WT animals). b Typical external pathology of a pancreatic tumor occurring in a tmKO animal. The inset shows an age-matched control pancreas. c Tumor incidence curves and Kaplan–Meier analysis comparing smKO (n = 13), dmKO (n = 29), and tmKO (n = 22) genotypes. Statistical analysis, done using the Prism Mantel–Cox test, revealed that SC- PaNEC incidences were significantly different across the experimental setups (p < 0.0001). Chi-Squared values and the Hazard ratios are listed in Supplementary Table 2D. d H&E stain of pancreatic tumors shows recurrent histological features of SC-PaNEC with necrotic foci (nec). The black arrow indicates nonneoplastic pancreatic tissue. The inset shows, under higher magnification, diffuse sheets of poorly dif- ferentiated cells with small blue round morphology, nuclear pleo- morphism, and hemorrhages (yellow arrows). e SC-PaNEC sections immunostained for pHH3 or PCNA, counterstained with Hoechst- 33342, reveal high proliferative capacity. f Laser-capture micro- dissection (LCM) of dmKO SC-PaNEC reveals chromosomal instability in tp53−/− tumors. In tp53+/del4 tumors the allelic ratio of the mutant allele remains around 50% (right graph). In contrast, two out of three tp53−/− tumors show substantial deviation from the expected allelic ratios (50% each variant) (left graph). g Percentage of mutant rbl1 and tp53 reads observed in LCM-derived SC-PaNECs sampled from either tmKO (left) or dmKO (right) animals. Red bullets and blue bullets demarcate SC-PaNECs with monoallelic inactivating tp53 mutations in the tmKO and dmKO setup, respectively. White scale bar is 500 μm, black scale bar is 50 μm.
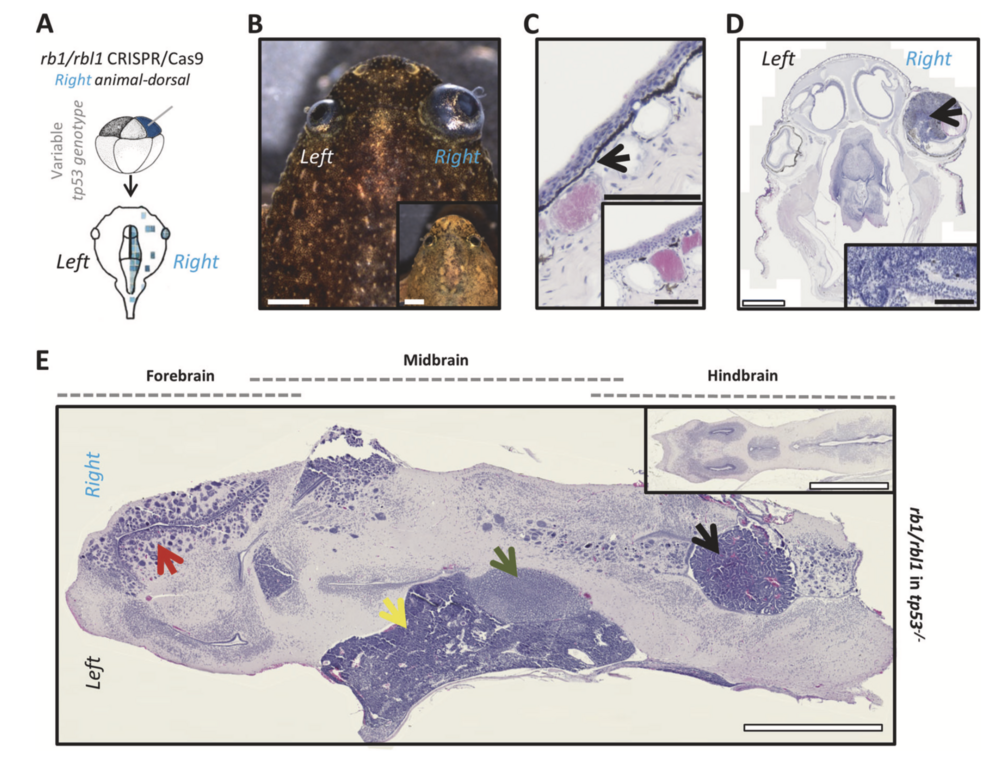
Fig. 3 Rb1 and rbl1 crispants (ectodermal targeted) develop reti- noblastoma, excessive black skin pigmentation and a spectrum of brain tumors. a CRISPR/Cas9 targeting of rb1 and rbl1 in the ectodermal lineage via unilateral injection of an animal-dorsal blas- tomere. b Unilateral ectodermal targeting of rb1/rbl1 CRISPR/Cas9, in tp53 wild-type, heterozygous, or compound heterozygous background, results in crispants developing externally visible retinoblastoma (75%; n = 16) and excessive skin pigmentation (44%; n = 16). Inset shows an animal from the same clutch without external symptoms. c H&E- stained section illustrating increased pigment deposition internal to the stratified epithelium (black arrow). Inset is an animal from the same clutch showing normal pigment deposition. d H&E-stained section of the animal shown in b. Unilateral retinoblastoma (black arrow) can clearly be distinguished. Inset shows higher magnification of the retinoblastoma illustrating the classic histopathological characteristics (Homer Wright rosettes). e H&E-stained horizontal brain section (anterior side of the animal to the left) revealing multiple neoplasms. Two distinct poorly differentiated, highly malignant, small blue round cell tumors (SBRCT) of which one is closely associated with the diencephalon (3th ventricle) and corresponds to a pinealoblastoma (green arrow). The other SBRCT has a deeper and more caudal localization towards the cerebellum, which is in line with the diagnosis of medulloblastoma (yellow arrow). Furthermore, a choroid plexus neoplasm can clearly be distinguished within an expanded ventricle (black arrow) and the forebrain harbors a glioblastoma lesion (red arrow). Inset shows the normal X. tropicalis brain architecture. White scale bar is 1 mm, black scale bar is 100 μM.
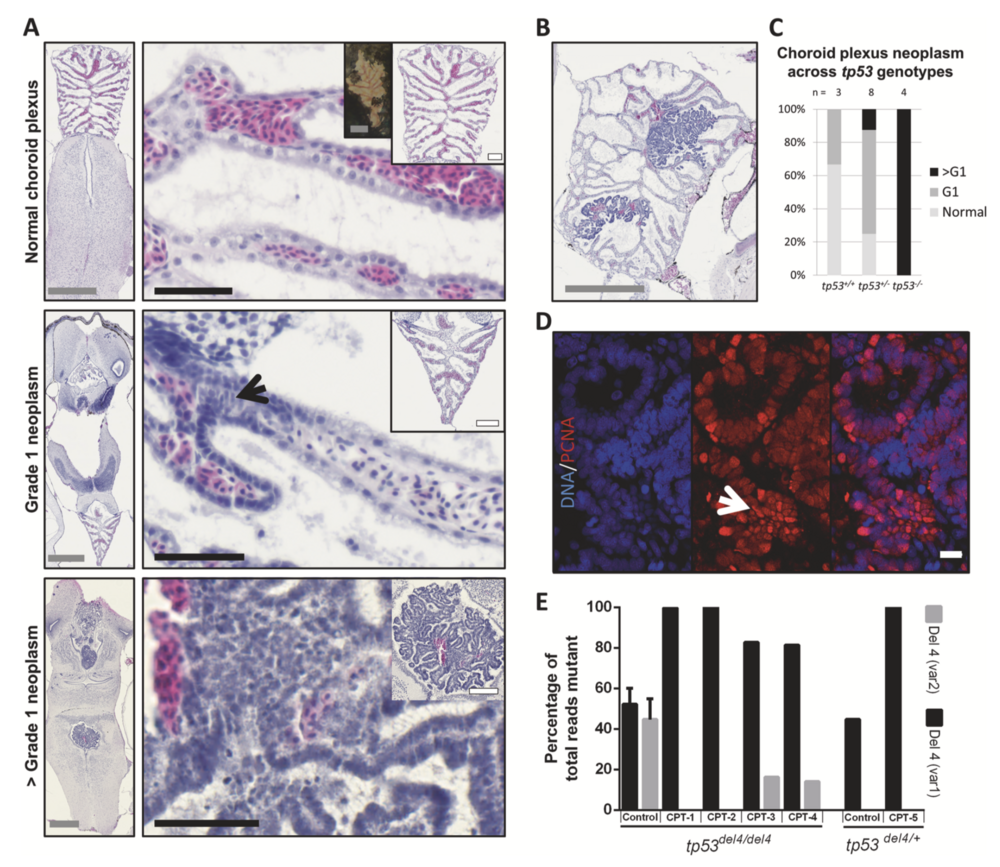
Fig. 4 Choroid plexus (CP) tumors arising in rb1/rbl1 crispants differ in grade according to their tp53 genotype. a H&E-stained sections of choroid plexuses representative for the CPT tumor grades observed within the clutch. (Top panels) Normal choroid plexus architecture consists out of a layer of ependymal cells, underlying capillaries and pia mater. Insets demonstrate both gross (left) and histological (right) normal CP architecture. Note that in Xenopus, the erythrocytes are nucleated. (Middle panels) WHO grade 1 (G1) CP neoplasm with cellular disarray, (pseudo)-stratification, atypia, and loss of polarization (black arrow). (Bottom panels) WHO > G1 CP neoplasm with pronounced atypia and aggressive growth character- istics. b Choroid plexus tumor in a tp53+/− animal showcasing clear continuity between normal CP and neoplastic tissue. c Association between tp53 genotype and grade of choroid plexus lesions (p < 0.01; Table S2I). d >G1 CP neoplasm immunostained for PCNA (red), counterstained with Hoechst-33342, reveals high proliferative capacity within highly atypical tumor areas (white arrow). e Laser-capture microdissection (LCM) of CP tumors (CPT) reveals chromosomal instability, as can be appreciated by the differential tp53 allelic ratios between control and CPT tissues. Left (tp53Δ4/Δ4) control bar show the average of LCM control tissues (n = 2). Right (tp53Δ4/+) control is the allelic ratio in spinal cord control tissue from the animal carrying CPT- 5. Gray scale bar is 500 μM, white scale bar is 100 μM, black scale bar is 50 μM. White scale bar in panel D is 10 μM.
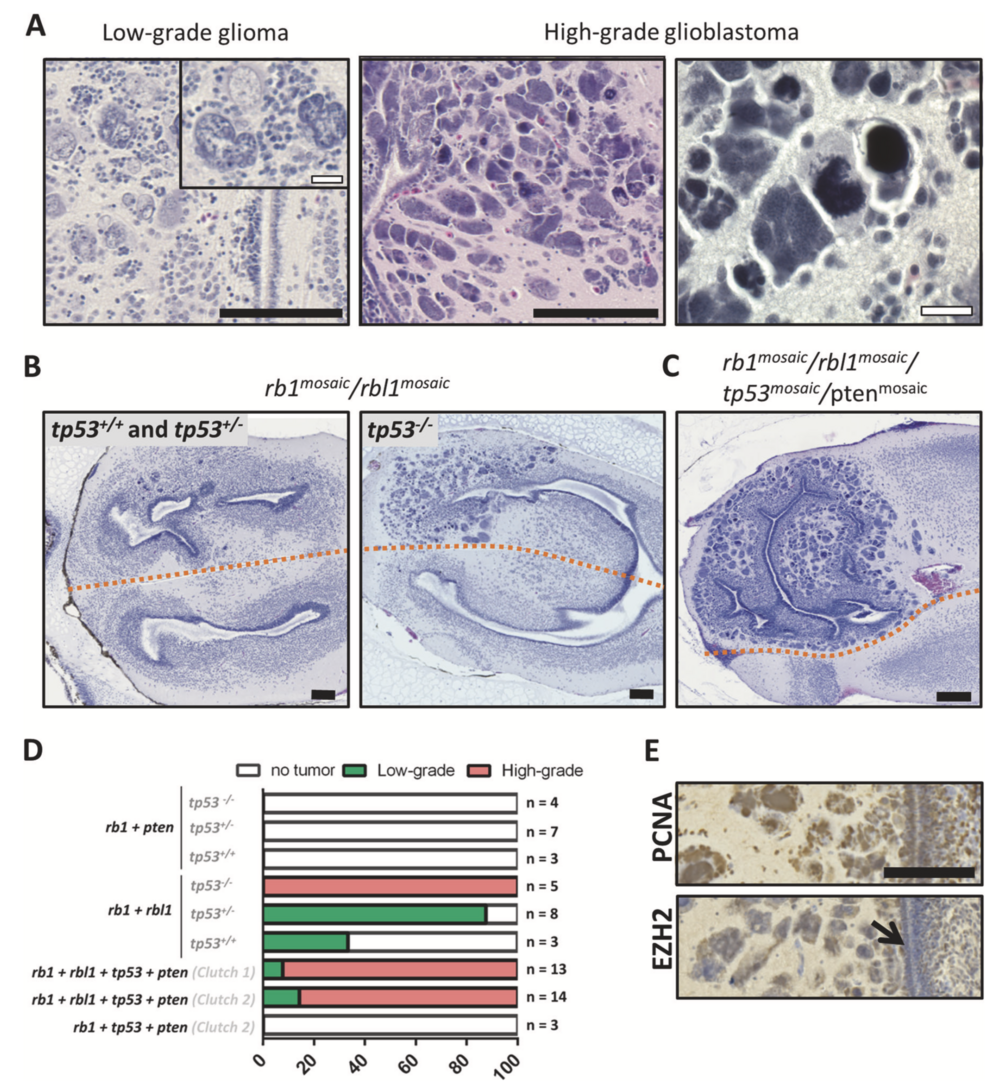
Fig. 5 Rbl1 functions as a tumor suppressor in glioblastoma, while tp53 inactivation underlies progression. a H&E-stained sections of Xenopus forebrains containing representative histopathology of low- grade glioma (left) and high-grade glioblastoma (middle and right) occurring across different experimental conditions. b (Left panel) Low-grade glioma with the occasional multinucleated giant cell in tp53 wild-type and heterozygous animals, targeted in the ectodermal lineage with rb1 and rbl1 CRISPR/Cas9. (Right panel) When similarly targeting rb1 and rbl1 in tp53 nullizygous animals, higher-grade glioblastoma lesions develop. c Quadruple multiplex CRISPR/Cas9- mediated targeting of rb1, rbl1, tp53, and pten in the ectodermal lineage leads to fully penetrant (100%; n = 13) development of high- grade glioblastoma lesions at day 42. d Bar plot representing the absence or presence of either low-grade glioma or high-grade glio- blastoma lesions across the performed experimental setups. Genes shown in bold were edited in the ectodermal lineage by CRISPR/Cas9, while tp53 genotypes, shown in dark gray, were germline inherited. e (top) PCNA immunostaining reveals high proliferative index in glio- blastoma cells. (bottom) EZH2 immunostaining reveals EZH2 expression in glioblastoma cells, but the absence of expression in the subventricular zone (black arrow). Black scale bar is 100μM and white scale bar is 20 μM.
Reprinted by permission from Springer Nature. Naert, T., Dimitrakopoulou, D., Tulkens, D. et al. RBL1 (p107) functions as tumor suppressor in glioblastoma and small-cell pancreatic neuroendocrine carcinoma in Xenopus tropicalis. Oncogene (2020). https://doi.org/10.1038/s41388-020-1173-z, Copyright 2020.
Last Updated: 2020-02-28