Bicc1 and Dicer regulate left-right patterning through post-transcriptional control of the Nodal inhibitor Dand5
Markus Maerker, Maike Getwan, Megan E. Dowdle, Jason C. McSheene, Vanessa Gonzalez, José L. Pelliccia, Danielle S. Hamilton, Valeria Yartseva, Charles Vejnar, Melanie Tingler, Katsura Minegishi, Philipp Vick, Antonio J. Giraldez, Hiroshi Hamada, Rebecca D. Burdine, Michael D. Sheets, Martin Blum & Axel Schweickert
Nature Communications volume 12, Article number: 5482 (2021)
Click here to view article at Nature Communications.
Click here to view article on Pubmed.
Click here to view article on Xenbase.
Abstract
Rotating cilia at the vertebrate left-right organizer (LRO) generate an asymmetric leftward flow, which is sensed by cells at the left LRO margin. Ciliary activity of the calcium channel Pkd2 is crucial for flow sensing. How this flow signal is further processed and relayed to the laterality-determining Nodal cascade in the left lateral plate mesoderm (LPM) is largely unknown. We previously showed that flow down-regulates mRNA expression of the Nodal inhibitor Dand5 in left sensory cells. De-repression of the co-expressed Nodal, complexed with the TGFß growth factor Gdf3, drives LPM Nodal cascade induction. Here, we show that post-transcriptional repression of dand5 is a central process in symmetry breaking of Xenopus, zebrafish and mouse. The RNA binding protein Bicc1 was identified as a post-transcriptional regulator of dand5 and gdf3 via their 3′-UTRs. Two distinct Bicc1 functions on dand5 mRNA were observed at pre- and post-flow stages, affecting mRNA stability or flow induced translational inhibition, respectively. To repress dand5, Bicc1 co-operates with Dicer1, placing both proteins in the process of flow sensing. Intriguingly, Bicc1 mediated translational repression of a dand5 3′-UTR mRNA reporter was responsive to pkd2, suggesting that a flow induced Pkd2 signal triggers Bicc1 mediated dand5 inhibition during symmetry breakage.
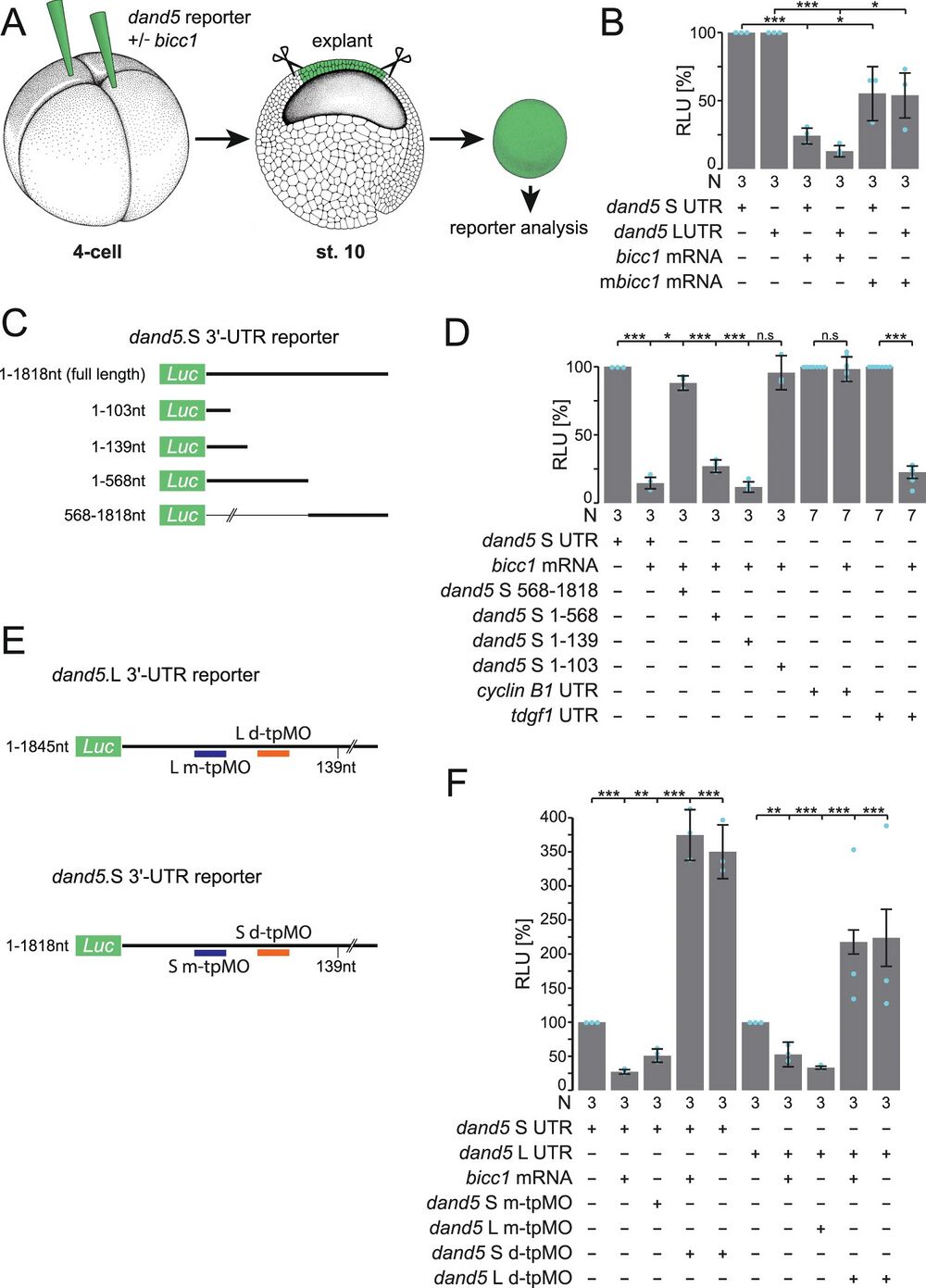
Fig. 1: Bicc1 represses dand5 mRNA translation via its proximal 3′-UTR.
A Schematic depiction of dand5 reporter assay. dand5 3′-UTR sequences fused to luciferase coding were injected either with or without bicc1 mRNA into the animal region of four-cell embryos. At st. 10, the animal cap region was excised and assayed for luciferase activity. Adapted from refs. 65 and 66. B Animal cap reporter assay following injections of dand5 S- or L 3′-UTRs alone or together with Xenopus (bicc1) or mouse Bicc1 (mbicc1) effector mRNAs. Note that both alloalleles were equally repressed. Note also that mbicc1 was efficient as a repressor as well. C Luciferase reporter constructs harboring different regions of the dand5 (S-allele) 3′-UTR. D Repression of translation is mediated through a proximal 139 nucleotides (nt) sequence element in the dand5 3′-UTR. E Schematic depiction of medial and distal target protector MOs (m-tpMOs or d-tpMOs) binding to the minimal Bicc1 responsive element (Bicc1RE) in the dand5 3′-UTR (L or S). F m- and d-tpMOs (0.4 or 0.5 pmol/embryo, respectively) interact differently with the luciferase reporter expression. m-tpMO blocked and d-tpMO boosted luciferase activity. Co-injection of d-tpMOs prevented Bicc1-dependent repression of the full-length dand5 reporters (L and S) and further enhanced their expressivity. N in B, D, and F represents the number of independent experiments. A pool of 10 animal caps was analyzed per experiment and treatment. Results from reporter mRNAs alone served as reference and were set to 100% RLU. Relative values of single experiments are depicted as blue dots. Data of at least three experiments are presented as mean value (bar) ±standard deviation (error bar, SD). Statistical analyses were done with a one-sided Student’s t test for two independent means (Bonferroni corrected) using the values of at least three individual experiments. p values, values for individual experiments, the mean values, and standard deviations are found in the source data file. n.s. not significant p > 0.05; * significant, p < 0.05; ** highly significant p < 0.01; ***, very highly significant p < 0.001; RLU relative luciferase units; Luc luciferase.
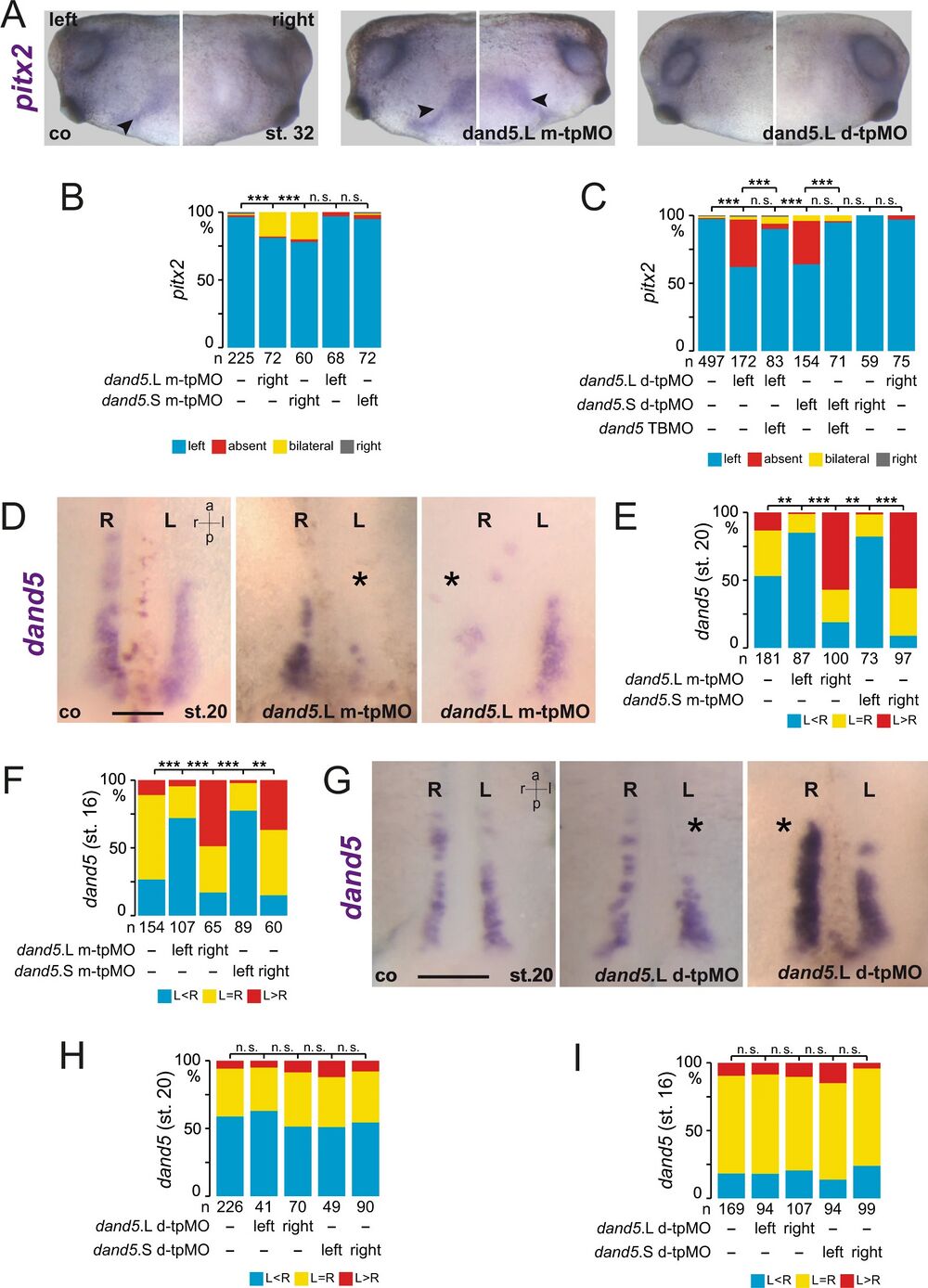
Fig. 2: Bicc1 responsive element (Bicc1RE) of the dand5 3′-UTR is required for LR asymmetry.
A Uninjected control (co), m-tpMO, or d-tpMO-injected embryos showed left, bilateral, or absent pitx2 expression, respectively. Lateral views (left and right) of embryos are presented. Arrowheads mark pitx2-positive lateral plate mesoderm. B Quantification of pitx2 results of m-tpMO-treated specimens. C Quantification of pitx2 asymmetry by d-tpMO injections. Note administration of dand5 TPMO together with d-tpMOs restored wt pitx2 expression. D Diminished dand5 mRNA expression by left-sided and right-sided m-tpMO injections compared with control. E Quantification of dand5 expression at post-flow stages (st.20) following m-tpMO treatment. F Quantification of dand5 expression in pre-flow specimens injected with m-tpMO. G Wildtype dand5 repression in control (co) and left- or right-sided d-tpMO injected specimens. H Quantification of dand5 asymmetry. Note flow-induced dand5 mRNA decay was observed in controls and following d-tpMO application. I Quantification of dand5 staining of pre-flow specimens (st.16) following d-tpMO injections. MO pmol/embryo: m-tpMO (L or S, 0.8); d-tpMO (L or S, 1). Asterisks in D and G mark injected side. Scale bars in D and G represent 100 µm. Numbers (n) in B, C, E, F, H, and I represent analyzed specimens from more than three independent experiments. Statistical analyses were done with one-sided Pearson’s chi-square test, which was adjusted for multiple comparisons by Bonferroni (B, C) or Bonferroni–Holm (E, F, H, I). p values and listing of individual experiments can be found in the source data file. n.s. not significant p > 0.05; **highly significant p < 0.01; ***very highly significant p < 0.001; st. stage; a anterior; l left; r right; p posterior.
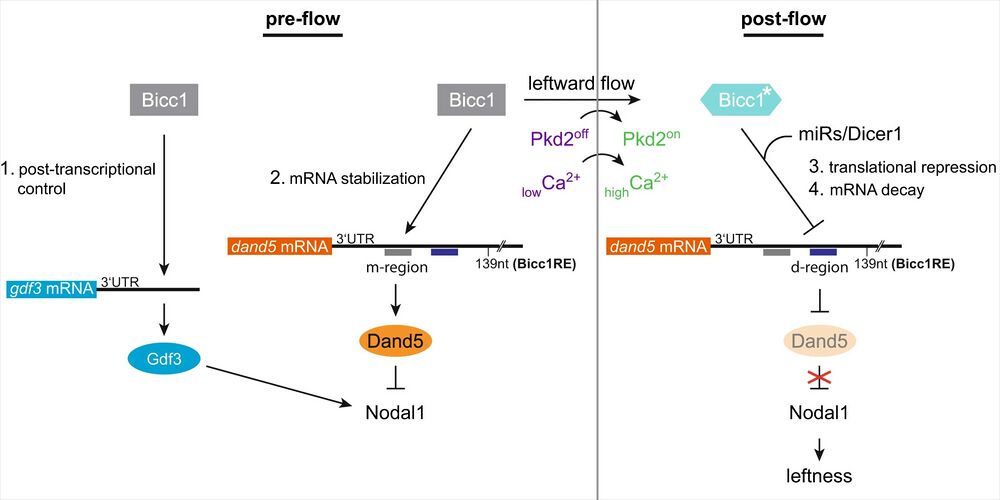
Fig. 8: Two modes of Bicc1-dependent post-transcriptional regulation of gdf3 and dand5 in flow sensor cells at the Xenopus left-right organizer.
In the early neurula pre-flow stages, Bicc1 has two functions. Bicc1 assures gdf3 mRNA translation and thereby indirectly ensures nodal1 transcription by Gdf3 signaling. Simultaneously Bicc1 mediates dand5 mRNA stability via the medial (m) sub-region of the Bicc1RE. Thus, Dand5 protein levels are sustained on both sides, keeping Nodal in tight repression. Leftward flow activates the Pkd2 channel in left flow sensor cells, resulting in an asymmetric calcium signal. In post-flow stages, a calcium-dependent mechanism activates/modifies Bicc1 to become a repressor of dand5 translation, which is relayed by the distal (d) sub-region of the Bicc1RE. Subsequently, dand5 mRNA gets degraded in a Dicer1 (miR) dependent manner. Attenuated Dand5 expression lifts repression of Nodal and defines leftness by induction of the LPM Nodal signaling cascade. For details, see text.
Adapted with permission from Springer Nature on behalf of Nature Communications: Maerker et al. (2021). Bicc1 and Dicer regulate left-right patterning through post-transcriptional control of the Nodal inhibitor Dand5. Nature Communications, 2021 Sep 16;12(1):5482. doi: 10.1038/s41467-021-25464-z.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2021-09-23