XB-IMG-148937
Xenbase Image ID: 148937
|
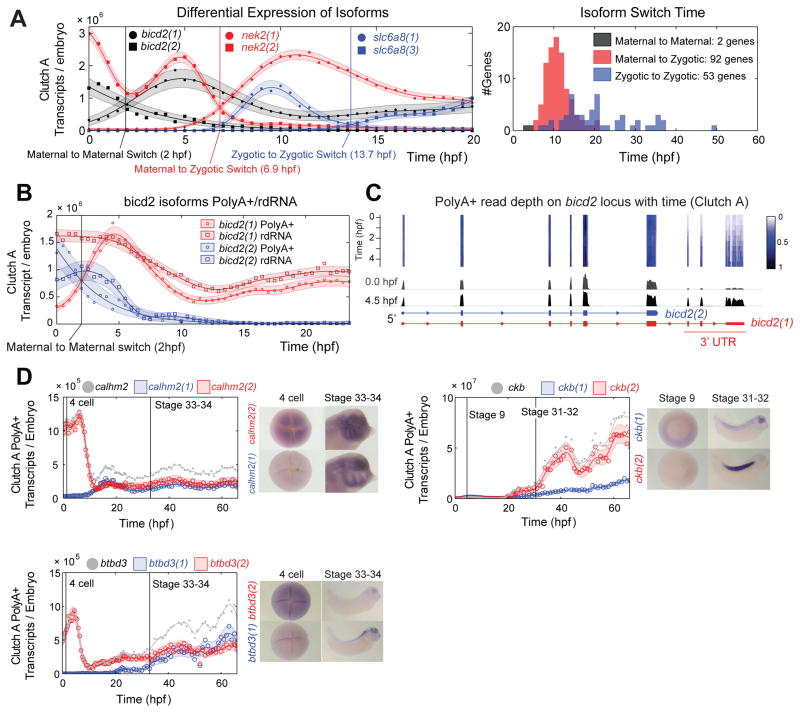
Isoform Differential Expression in Clutch A, see also Fig S5
A â Isoform switching event examples: maternal to maternal; maternal to zygotic; and zygotic to zygotic switching events (left). Histogram of isoform switching events by category and time (right).
B â bicd2 isoforms switch in PolyA+ data (circles), and do not switch in rdRNA data (squares) indicating differential polyadenylation. PolyA+ and rdRNA abundances agree within uncertainty of the absolute normalization (Supplemental Experimental Procedures).
C â Normalized read depth on bicd2 locus in Clutch A polyA+ between 0â4.5 hpf. Note temporal dynamics shared by the final three bicd2(1) exons. Heatmap and depth of pile ups corrected for changing total mRNA.
D â Spatial expression of three genes with isoforms with differential dynamics. All isoforms show different temporal and spatial expression domains. Grey lines and points mark total expression for each gene (sum of the red and blue isoforms). Image published in: Owens ND et al. (2016) Copyright © 2016. Image reproduced with permission of the Publisher and the copyright holder. This is an Open Access article distributed under the terms of the Creative Commons Attribution License.
Image source: Published Larger Image Printer Friendly View |
