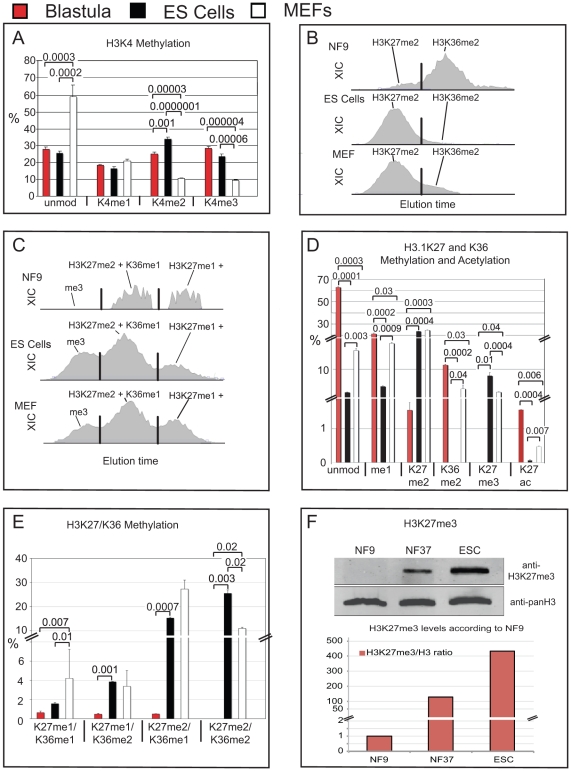
Figure 5. Comparison of opposing histone modifications on histone H3 from Xenopus blastulae, murine ES cells and MEFs.A) Bar-Charts showing H3K4 methylation states of X.laevis blastulae, GS-1 ES cells and mouse embryo fibroblasts. Data obtained by MALDI-TOF mass spectrometry, error bars indicate SD of three independent biological replicates. B) XIC profiles of K27me2 and K36me2 modification states of the H3 27–40 peptide of X.laevis blastulae, murine ES Cells and MEFs on a C18 micro-column on a reversed-phase HPLC during Orbi-Trap on-line mass spectrometry. C) XIC profiles for K27/K36me3, K27me2+K36me1 and K27me1+K36me2 of the H3 27–40 peptides from the same samples. In B and C, x-axis gives relative elution times of peptides, the y-axis shows their intensity according to the ion current of the quadrupole Mass Spectrometer. In B and C, X-axis represents retention time, y-axis the intensity of ion currents of the quadrupole Orbi-Trap mass spectrometer. Peak separation was called by the ICIS peak detection algorithm program (Thermo), indicated here by vertical black lines. D) Bar-Chart of mutually exclusive H3K27 and K36 methyl states. E) Bar-Chart of peptides with combinatorial H3K27 and K36 methylation. D and E - data from Orbi-Trap Mass Spectrometry, error bars indicate SEM of two independent biological replicates. F) Immunoblotting for total histone H3 and histone H3 trimethylated at lysine 27. Upper panel – western blot Odyssey infrared imaging signals; lower panel – bar chart showing the abundance of H3K27me3 levels relative to total histone H3 protein in Blastulae (NF9), Tadpoles (NF37) and murine GS-1 ES cells. Abbr.: unmod = unmodified peptide, Kme1 = mono-methylated lysine residue, Kme2 = di-methylated lysine residue, Kme3 = tri-methylated lysine residue. Where applicable, p values are given by numbers above brackets to indicate significant differences in histone modifications between samples.
Image published in: Schneider TD et al. (2011)
Schneider et al. Creative Commons Attribution license
Permanent Image Page
Printer Friendly View
XB-IMG-126247