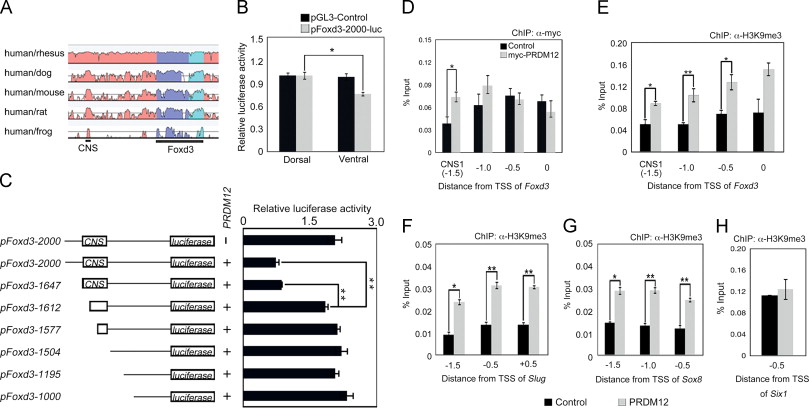
Fig. 3. PRDM12 promotes trimethylation of H3K9 on Foxd3 promoter regions through binding the CNS. (A) VISTA view of the conserved sequence domains in the genomic region containing the Foxd3 gene. Colored peaks indicate regions of at least 100 bp and with 60% similarity [dark blue, exons; light blue, untranslated regions (UTRs); pink, noncoding regions]. One conserved noncoding sequence (CNS) is shown. (B and C) Luciferase assays using the indicated Foxd3 promoter region. Expression levels were normalized to Renilla luciferase. Error bars represent ±S.D. (n=3). (B) Luciferase assays on stage 20 embryos that were co-injected at the four-cell stage with pGL3-Control or pFoxd3-2000-luc and pGL-TK (Renilla luciferase vector) into the dorsal or ventral side. (C) The indicated luciferase reporter vector and PRDM12 mRNA were co-injected into the dorsal side of four-cell-stage embryos, which were harvested after injection and subjected to relative luciferase activity measurement. (DâH) ChIP-qPCR analysis using the Chd and Wnt8 mRNA-injected animal cap cells co-injected with or without myc-PRDM12 mRNA. The vertical axes represent the percentage input (ChIP enriched/input) and the horizontal axes represent the approximate distance from the transcriptional start site (TSS) of each gene. ChIP-qPCR analyses were performed with an anti-myc antibody (D) or anti-H3K9me3 antibody (EâH). The Six1 promoter region (â0.5 kbp from the TSS) was used as a negative control. Error bars indicate ±S.E. (n=3). Differences were considered to be statistically significant at pâ¤0.05(*) or pâ¤0.01(**) by Studentâs t-test.
Image published in: Matsukawa S et al. (2015)
Copyright © 2015. Image reproduced with permission of the Publisher, Elsevier B. V.
Permanent Image Page
Printer Friendly View
XB-IMG-135994