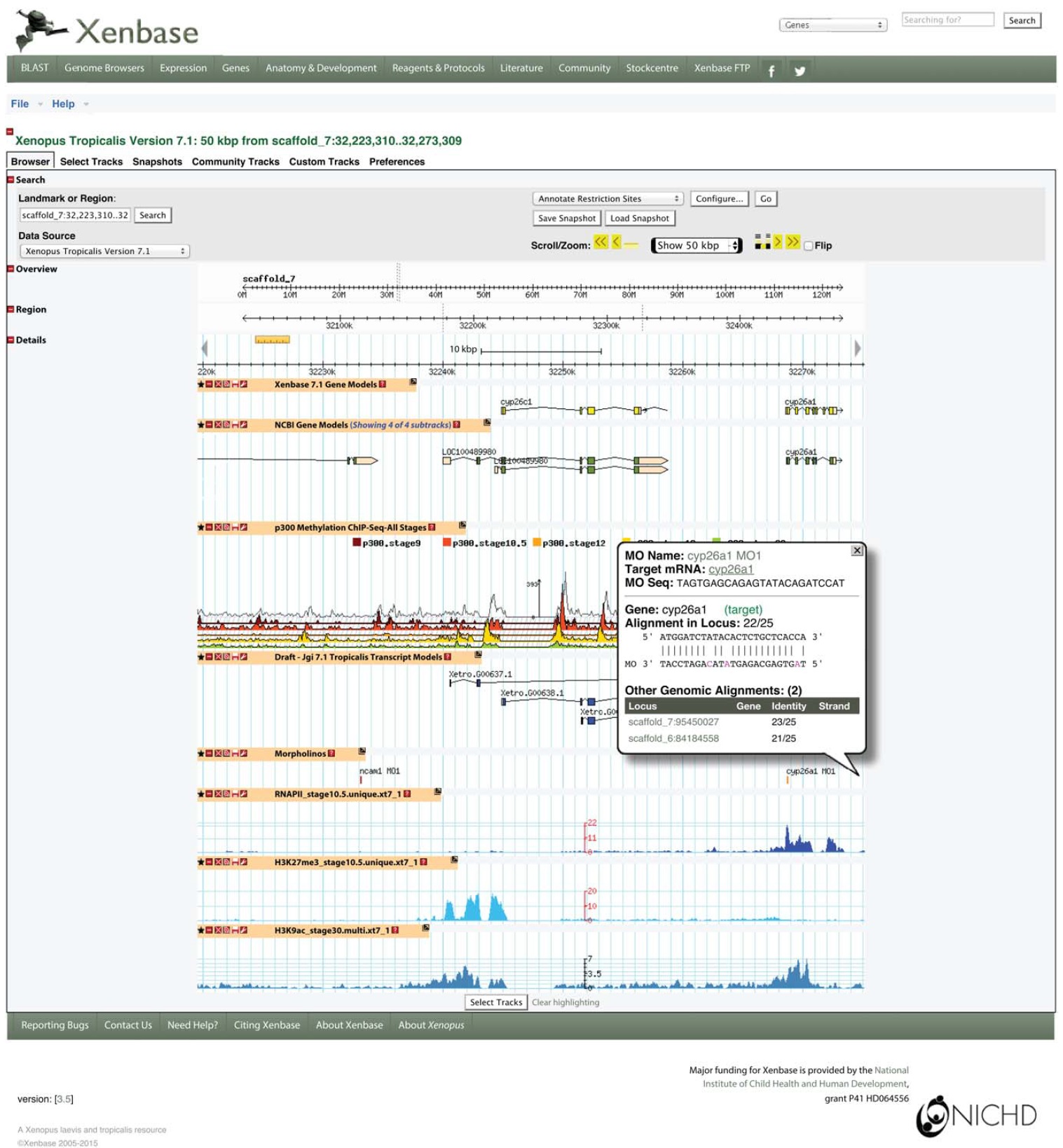
FIG. 3. The Xenbase browser can load three different builds of both X. tropicalis (4.1, 7.1, and 8.0) and X. laevis (6.0, 7.2 [both J strain], and WT1.0). Multiple gene models, their annotations, and a variety of epigenomic and ancillary data are available for most genome builds. Many tracks launch pop-up windows with further information. In this view, a user has clicked on the morpholino feature labeled âcyp26a1 MO1.â This reagent was designed against X. laevis cyp26a; so, a number of mismatches are present in the alignment to the active view, which is X. tropicalis build 7.1. The stacked ChIP-seq tracks for p300 are displayed using Topoview (available at: http://flybase.org/static_ pages/docs/software/topoview.html).
Image published in: James-Zorn C et al. (2015)
Copyright © 2015. Image reproduced with permission of the Publisher, John Wiley & Sons.
Permanent Image Page
Printer Friendly View
XB-IMG-145537