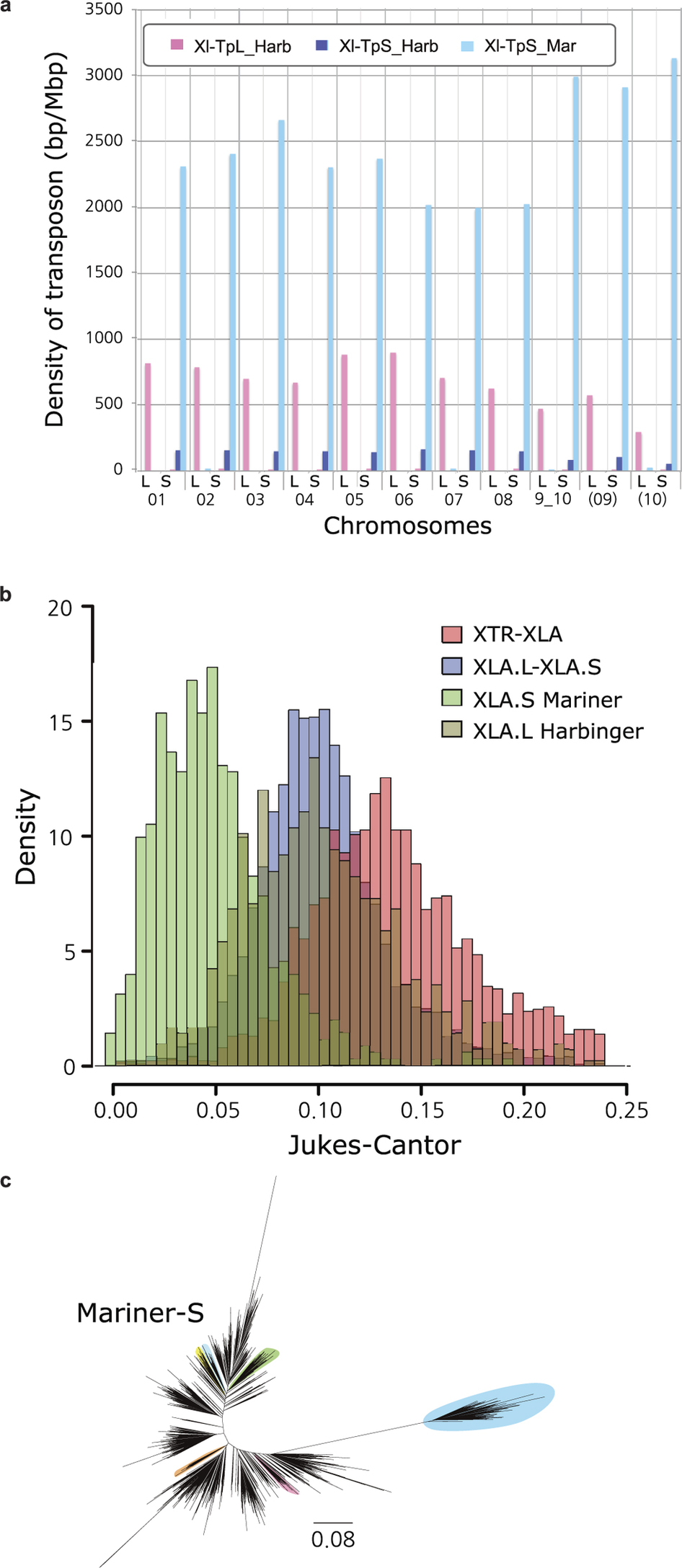
EDF 3. TRANSPOSONS Density of the subgenome specific transposons on each chromosome (coverage length of transposable element [bp]/chromosome length [Mbp]). The coverage lengths of transposons were calculated from the results of BLASTN search (E-value cutoff 1E-5) using the consensus sequences as queries.Jukes-Cantor distances across non-CpG sites, corrected as in Supplemental Note 7.5. Distances between X. tropicalis and X. laevis transposons consensus sequences are shown. The X. laevis-specific transposon differences are each individual transposon sequence against the consensus sequence for that subfamily.Phylogenetic tree of Xl-TpS_Mar transposon expansions in the X. laevis genome, built using Jukes-Cantor corrected distances (Supplemental Note 7.5). Sub-clusters with enough members to determine accurate timings are highlighted. The scale bar represents the corrected Jukes-Cantor distance.
Image published in: Session AM et al. (2016)
Image downloaded from an Open Access article in PubMed Central. Image reproduced on Xenbase with permission of the publisher and the copyright holder.
Permanent Image Page
Printer Friendly View
XB-IMG-153333