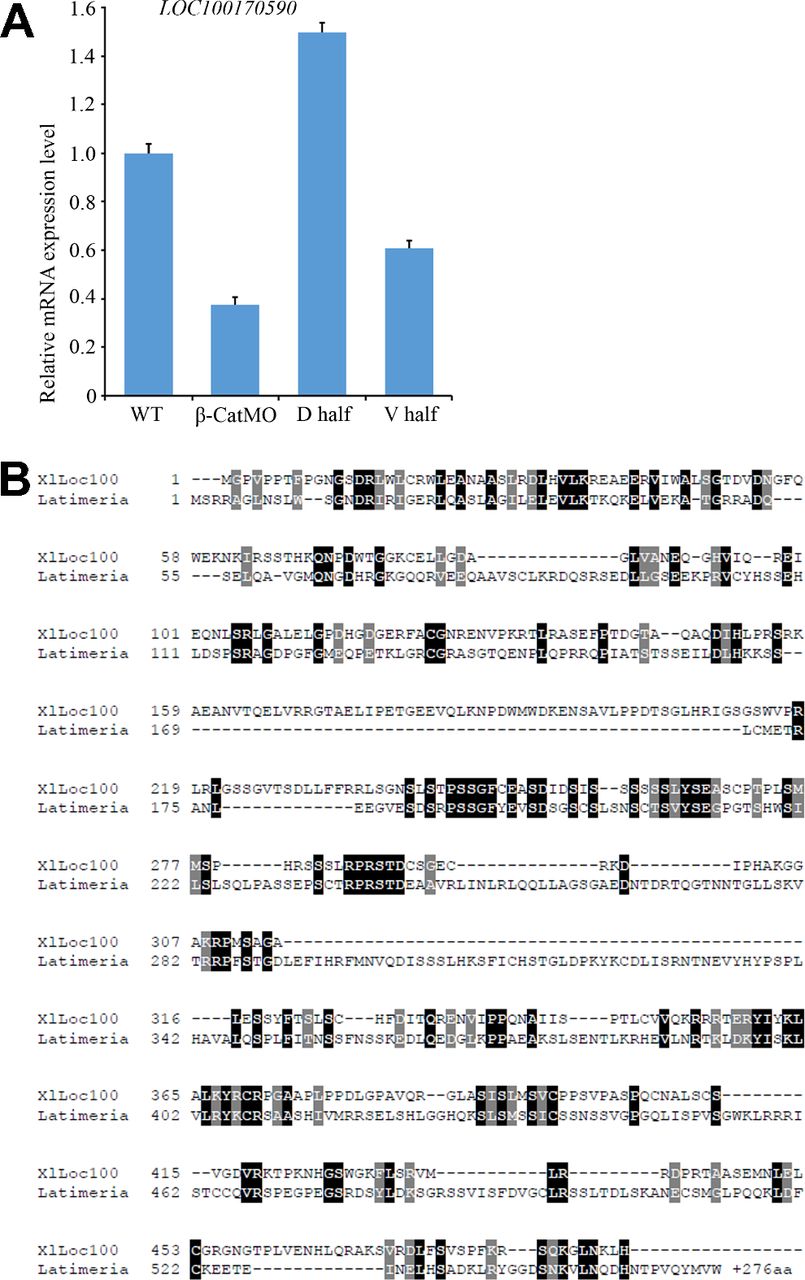
Fig. S2. qRT-PCR validation and sequence homology of LOC100170590. (A) β-catenin–dependent regulation and D-V enrichment of LOC100170590 was validated by qRT-PCR analysis. β-CatMO decreased LOC100170590 mRNA levels compared with uninjected WT controls (WT), and expression in dorsal half-embryos (D half) was significantly higher than in ventral half-embryos (V half). (B) Amino acid sequence comparison between Xenopus laevis Loc100170590 and Latimeria chalumnae XP_005988953.1 (a coelacanth fish) revealed a 27% degree of identity. The coelacanth gene has been annotated as possible Dapper homolog-like 2, but XlLoc100170590 lacks critical Dapper-like features, such as a PDZ domain and a leucine zipper. The sequence alignment was generated by using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) followed by BoxShade (embnet.vital-it.ch/software/BOX_form.html). Black and gray shaded boxes indicate identical and similar amino acid residues, respectively. There were no significant homologies with mammalian genomes.
Image published in: Ding Y et al. (2017)
Copyright © 2017. Image reproduced with permission of the publisher and the copyright holder. This is an Open Access article distributed under the terms of the Creative Commons Attribution License.
Permanent Image Page
Printer Friendly View
XB-IMG-170549