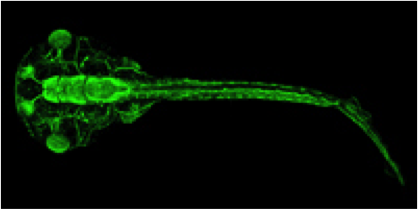
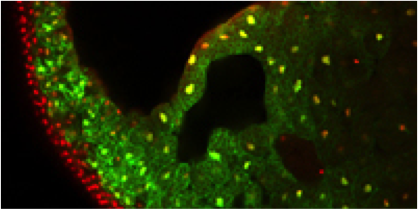
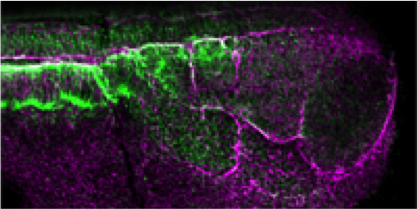
Xenopus is an essential vertebrate model system for
biomedical research
- Share 83% human disease genes
- Ease of genomic manipulation
- Large eggs and embryos with rapid external development
- Ease of housing
- Produce hardy eggs year-round
- Learn
more about Xenopus

X. tropicalis
Xenopus tropicalis (the western clawed frog)
The tropical clawed frog,
Xenopus
tropicalis [Pipidae], is native to several countries of
southwestern Africa including notably Cote d'Ivoire, Nigeria,
Senegal, and Sierra Leone. Sometimes also called the Western
clawed frog and previously
Silurana
tropicalis,
X. tropicalis is fully
aquatic like
X. laevis and inhabits
mostly rainforests in West Africa. It is a smaller species than
X. laevis, with adult males measuring
3-4cm and females 4.5-5.5cm from snout to vent. Typical of
Xenopus species, it has a flattened body with a mottled or
blotchy dark gray, green and/or brown dorsal skin, with a pale or
unpigmented belly.
X. tropicalis is
another Xenopus species widely used in biological and biomedical
research. While there are fewer eggs per brood in
X. tropicalis than in
X. laevis, the former may be better
suited for certain genetic studies due to its simpler, diploid
genome.
X. tropicalis was the first
frog to have it's genome sequenced in 2010. The
X. tropicalis genome, currently available
in annotation v10.0 on Xenbase, has considerable sequence and
gene order conservation with other tetrapod vertebrates including
mammals, birds, reptiles and fish.
Learn more about the genomics of
Xenopus
.

X. laevis
Xenopus laevis (the African clawed frog)
The African clawed frog, Xenopus
laevis [Pipidae], is endemic to the African Rift Valley and
southern Africa with introduced populations in Europe, Asia and
North America. It is a large, fully aquatic species with a
flattened appearance and pronounced sexual dimorphism; Males are
generally smaller (4.5-10cm) than females (6-15cm). The
forelimbs are held extended, while hindlimbs are large muscular
with fully webbed toes. Both hands and feet have distinct black
toe tips resembling claws. Adults have dorsal skin patterns of
blotchy green, gray and brown with lighter colored bellies,
while albino varieties are also common in captivity. It is the
most widely used Xenopus species in biomedical research,
with a long history of use in embryology, cell biology and
developmental biology. The genome of X.
laevis, sequenced in 2016, is allotetraploid due to a
hybridization event that occurred 17–18 MYA between two extinct
diploid ancestors. X. laevis thus carries 2 subgenomes, referred
to as the ‘Long’ and ‘Short’ chromosomes. We assign a ‘.L’ or
‘.S’ suffix respectively to gene symbols to indicate to which
ancestral genome they belong. It is estimated that X. tropicalis
and X. laevis, diverged
approximately 48 MYA. The X. laevis
genome annotation v10.1 is available on Xenbase and other
resources.
Learn more about the genomics of
Xenopus
.

Nanorana parkeri
Nanorana parkeri (the Tibetan frog)
The Tibetan frog,
Nanorana parkeri
[Dicroglossidae], is endemic to the Tibetan Plateau and the
Himalayan mountain regions in China, Nepal, India and Bhutan. It
is commonly known as Parker’s slow frog, the mountain slow frog,
the Himalaya frog, or the Xizang Plateau frog. Adults have olive
green dorsal skin with brown or black stripes, including a
characteristic pair of stripes from the snout to each side of the
face. These frogs have adaptations to high elevations that
include changes to the cardiovascular system and tolerance to UV
radiation and hypoxia. They breed naturally in high altitude
marshes and streams and can also be found in highland forests,
grasslands and rivers.
The genome of N. parkeri was
sequenced in 2015 and has current assembly v1.0 available on
Xenbase via these links below:
JBrowse,
BLAST,
Download

Hymenochirus boettgeri
Hymenochirus boettgeri (the Congo dwarf clawed frog)
The Congo dwarf clawed frog,
Hymenochirus
boettgeri [Pipidae], is found in the Democratic Republic of
Congo, the Central African Republic, Nigeria, Cameroon, Gabon,
Equatorial Guinea. Previously known as
Xenopus
boettgeri, or otherwise commonly as the Zaire dwarf clawed frog
or the dwarf African clawed frog, it is now considered the
closest outgroup for the
Xenopus genus. Smaller than other
Xenopus frogs, they have long and thin legs, clawed hind
feet, tapered heads and gray-brown dorsal skin with small dark
spots. The natural habitat is slow moving or still waters in
rainforest lowlands, although it is a common species in the
aquarium trade worldwide. The genome of
H.
boettgeri was sequenced in 2021 and the current assembly v1.0 is
available on Xenbase via these links below:
JBrowse,
BLAST,
Download

Ambystoma mexicanum
Ambystoma mexicanum (the Mexican axolotl)
The Mexican axolotl,
Ambystoma
mexicanum [Ambystomatidae], is endemic to only two lakes, Lake
Xochimilco and Lake Chalco, near Mexico City, Mexico. It is a
large species of neotenic salamander reaching lengths of 30cm
with short limbs and protruding gills. The axolotl’s skin is dark
in the wild however an albino variety is commonly bred in
captivity for the aquarium trade. Prominently known for its
neoteny, it remains in its larval body form into adulthood,
however it can metamorphose into the Mexican salamander in
conditions of environmental desiccation. The axolotl is an
emerging model organism notably for the study of tissue
regeneration and repair, neurulation, genomics, eye and heart
development and other topics.
Sequenced in 2021, the axolotl genome assembly v6.0
is available on Xenbase via these links below:
JBrowse,
BLAST,
Download

Lithobates catesbeianus
Lithobates catesbeianus (the American bullfrog)
The American bullfrog, Rana
(Lithobates) catesbeianus [Ranidae], is native to Canada,
Mexico and the United States, however is invasive to several
countries in Europe, Asia and South America. Previously known as
Rana catesbeiana, it was reassigned to the genus Lithobates in
2006 and has since been argued that Lithobates may best be
considered a subgenus of the genus Rana. It is also known by its
homotypic synonym as Aquarana catesbeiana.
These are the largest frogs in North America, with the larger
females growing up to 180mm in length from snout to vent. Males
have large, defining tympanums wider in diameter than the eyes.
Adults have green dorsal skin with a dark, netlike pattern on
top, however skin colour varies by region. They live and breed
in vegetation-covered shallow waters of lakes and marshes.
Bullfrogs are territorial of breeding sites and prey on any
animal smaller than themselves including other amphibians,
insects, fish, mice and crayfish. They are an important source
of food consumption and are used for pest control in certain
regions. The genome of L.
catesbeianus was sequenced in 2017 with the v2.1 assembly
currently available on Xenbase via these links below:
JBrowse, BLAST,
Download

The Xenopus model organism knowledgebase
Xenbase is a web-accessible resource that integrates all the
diverse biological, genomic, genotype and phenotype data
available from
Xenopus research.
Learn
more about Xenbase.

Gene Expression Datasets
Xenopus Data Sources