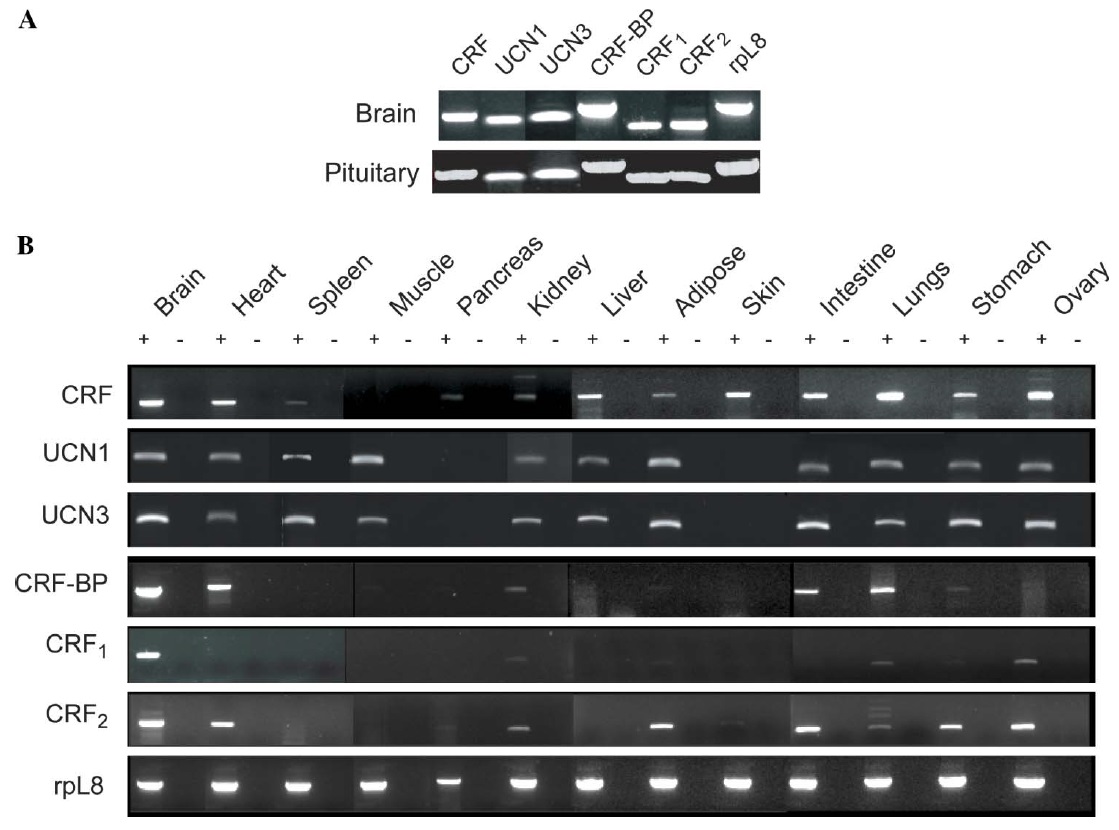
Fig. 2. Tissue distribution of mRNAs for corticotropin-releasing factor system components in juvenile Xenopus laevis tissues analyzed by RT-PCR. RNA isolation and RT-PCR is described by Boorse et al. (2005a) and the PCR primer sequences are given in Table 1. (A) Expression analysis in brain and pituitary. (B) Expression analysis in diverse peripheral tissues. Corticotropin-releasing factor (CRF), urocortin 1 (UCN1), urocortin 3 (UCN3), CRF binding protein (CRF-BP), CRF receptor 1 (CRF1) and CRF receptor 2 (CRF2). Ribosomal L8 (rpL8) was used as a housekeeping gene to control for RNA quality and loading. Shown are representative ethidium bromide-stained gels of RT-PCR products; RNA was harvested from tissues of three females and similar results were obtained with each. PCR reactions conducted on reverse transcription reactions in which the reverse transcriptase was omitted (minus RT reactions) produced no bands, thus conWrming the absence of genomic DNA contamination + and ¡ on the Wgure. RT-PCR reactions conducted on RNA isolated from whole blood or packed blood cells produced no bands, which shows that the positive results that we obtained in other tissues are not due to contamination from blood cells. However, we cannot rule out the possibility that some blood cell types express CRF system signaling components but their abundance was below the level of detection of our assay.
Image published in: Boorse GC and Denver RJ (2006)
Copyright © 2006. Image reproduced with permission of the Publisher.
| Gene | Synonyms | Species | Stage(s) | Tissue |
|---|---|---|---|---|
| crh.L | corticoliberin, corticotropin-releasing factor, crh-a, crh-b | X. laevis | Sometime during NF stage 1 to NF stage 66 | |
| ucn3.L | LOC445842 | X. laevis | Sometime during NF stage 1 to NF stage 66 | |
| crhr1.2.S | CRF1, crf-r1, crfr1 | X. laevis | Sometime during NF stage 1 to NF stage 66 | |
| crhr2.S | CRF2, crf-r2, crfr2 | X. laevis | Sometime during NF stage 1 to NF stage 66 | |
| crhbp.S | CRF-BP, crfbp, CRH-BP, gene E | X. laevis | Sometime during NF stage 1 to NF stage 66 | |
| ucn1.L | X. laevis | Sometime during NF stage 1 to NF stage 66 |
Image source: Published
Permanent Image Page
Printer Friendly View
XB-IMG-153343