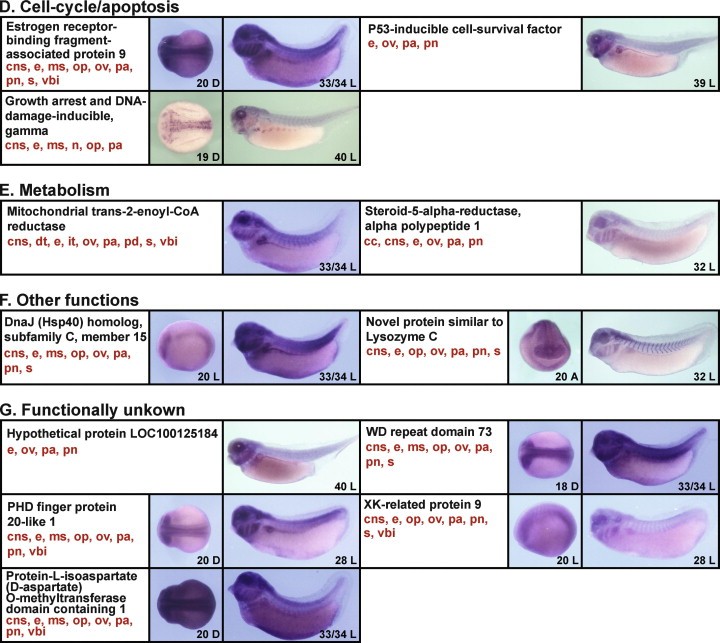
Fig. 4. The expression patterns of clones identified by the functional screen, examined by whole mount in situ hybridization using X. tropicalis embryos. Twenty-nine out of the 31 positive clones identified in the screen displayed identifiable in situ patterns of expression. The clones were divided into each of seven functional groups on the basis of their predicted/established function (A, transcription; B, cell signaling; C, post-transcription/translation; D, cell-cycle/apoptosis; E, metabolism; F, other functions; G, functionally unknown). The images are ordered by gene names (black). The main expression domains of each clone at all stages analyzed are indicated in red, although not all of these domains are represented in the photographs presented (cc, ciliated cells; cns, central nervous system; dt, distal tubules; e, eye; it, intermediate tubules; ms, migrating somite; n, primary neuron; ns, nephrostomes; op, olfactory pit; ov, otic vesicle; pa, pharyngeal arches; pcv, posterior cardinal veins; pn, pronephros; s, somite; vbi, ventral blood island). The numbers in each photograph indicate the developmental stage of embryos shown. Other abbreviations are: A, anterior view (dorsal is up); D, dorsal view (anterior is left); L, lateral view (anterior is left, dorsal is up).
Image published in: Kyuno J et al. (2008)
Copyright © 2008. Image reproduced with permission of the Publisher, Elsevier B. V.
| Gene | Synonyms | Species | Stage(s) | Tissue |
|---|---|---|---|---|
| wdr73 | X. tropicalis | Sometime during NF stage 18 to NF stage 33 and 34 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch pronephric kidney | |
| gadd45g | xgadd45g, XGadd45-gamma | X. tropicalis | Sometime during NF stage 19 to NF stage 40 | central nervous system eye somite neuron olfactory pit pharyngeal arch |
| phf20l1 | X. tropicalis | Sometime during NF stage 20 to NF stage 28 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch ventral blood island pronephric mesenchyme | |
| xkr9 | XK-related protein 9, Xrg9 | X. tropicalis | Sometime during NF stage 20 to NF stage 28 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch ventral blood island pronephric mesenchyme |
| spaca5l.2 | lyc5, lyzl5, sllp2 | X. tropicalis | Sometime during NF stage 20 to NF stage 32 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch pronephric kidney |
| ebag9 | X. tropicalis | Sometime during NF stage 20 to NF stage 33 and 34 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch pronephric kidney ventral blood island | |
| dnajc15 | X. tropicalis | Sometime during NF stage 20 to NF stage 33 and 34 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch pronephric kidney | |
| pcmtd1 | LOC108695289 | X. tropicalis | Sometime during NF stage 20 to NF stage 33 and 34 | central nervous system eye somite olfactory pit otic vesicle pharyngeal arch pronephric kidney ventral blood island |
| srd5a1 | SRD5alpha1 | X. tropicalis | Throughout NF stage 32 | ciliated epidermal cell central nervous system eye otic vesicle pharyngeal arch pronephric kidney |
| mecr | X. tropicalis | Throughout NF stage 33 and 34 | central nervous system pronephric nephron eye somite otic vesicle pharyngeal arch pronephric kidney ventral blood island | |
| triap1 | P53csv, p53-inducible cell-survival factor, triap1-a, triap1-b | X. tropicalis | Throughout NF stage 39 | eye otic vesicle pharyngeal arch pronephric kidney |
| vwa8 | X. tropicalis | Throughout NF stage 40 | eye otic vesicle pharyngeal arch pronephric kidney |
Image source: Published
Permanent Image Page
Printer Friendly View
XB-IMG-39363