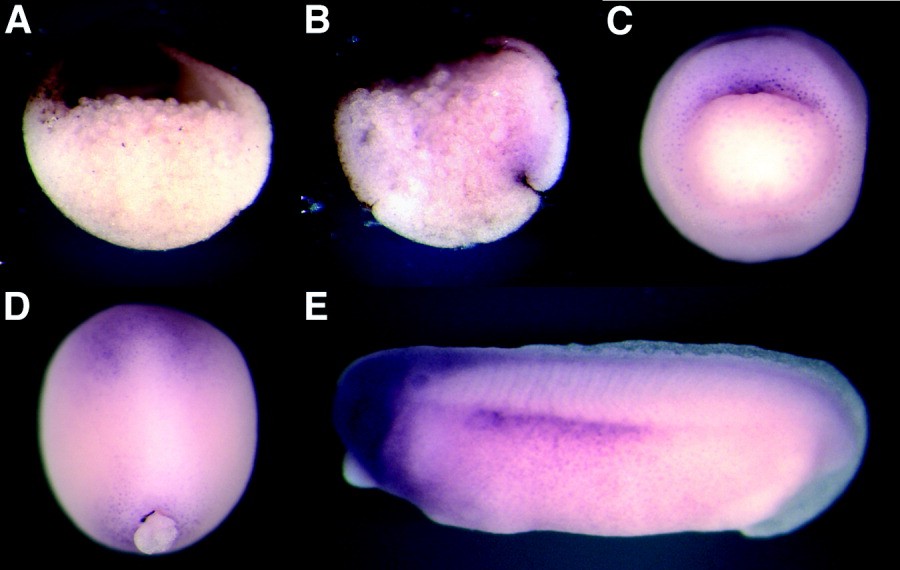
Figure 3. Spatial distribution of FRIED transcripts in Xenopus embryos. Wholemount in situ hybridization analysis has been carried out with digoxygenin-labeled FRIED anti-sense RNA probes with embryos at different developmental stages. A: Stage 10, animal pole is at the top, dorsal is to the right. FRIED RNA is detectable in animal pole ectoderm of a bisected embryo. B,C: At stage 10+, FRIED transcripts are visible in the dorsal blastopore lip. A,B: Saggital view. C: Vegetal pole view. D: Stage 14, dorsal view, anterior is at the top. FRIED is expressed in anterior ectoderm area and around closed blastopore at the beginning of neurulation. E: Tailbud stage, anterior is to the left. FRIED transcripts are clearly detectable in the brain and spinal cord, eyes and otic vesicles, and in lateral plate mesoderm. A control RNA probe did not produce similar staining patterns (data not shown).
Image published in: Itoh K et al. (2005)
Copyright © 2005. Image reproduced with permission of the Publisher, John Wiley & Sons.
| Gene | Synonyms | Species | Stage(s) | Tissue |
|---|---|---|---|---|
| ptpn13.L | fap-1, fried, Frizzled interaction and ectoderm defects, pnp1, ptp1e, ptp-bas, ptp-bl, ptpl1, ptple | X. laevis | Throughout NF stage 10 | animal hemisphere ectoderm upper blastopore lip |
| ptpn13.L | fap-1, fried, Frizzled interaction and ectoderm defects, pnp1, ptp1e, ptp-bas, ptp-bl, ptpl1, ptple | X. laevis | Throughout NF stage 14 | ectoderm anterior neural plate circumblastoporal collar |
| ptpn13.L | fap-1, fried, Frizzled interaction and ectoderm defects, pnp1, ptp1e, ptp-bas, ptp-bl, ptpl1, ptple | X. laevis | Throughout NF stage 28 | brain eye otic vesicle spinal cord lateral plate mesoderm |
Image source: Published
Permanent Image Page
Printer Friendly View
XB-IMG-44476