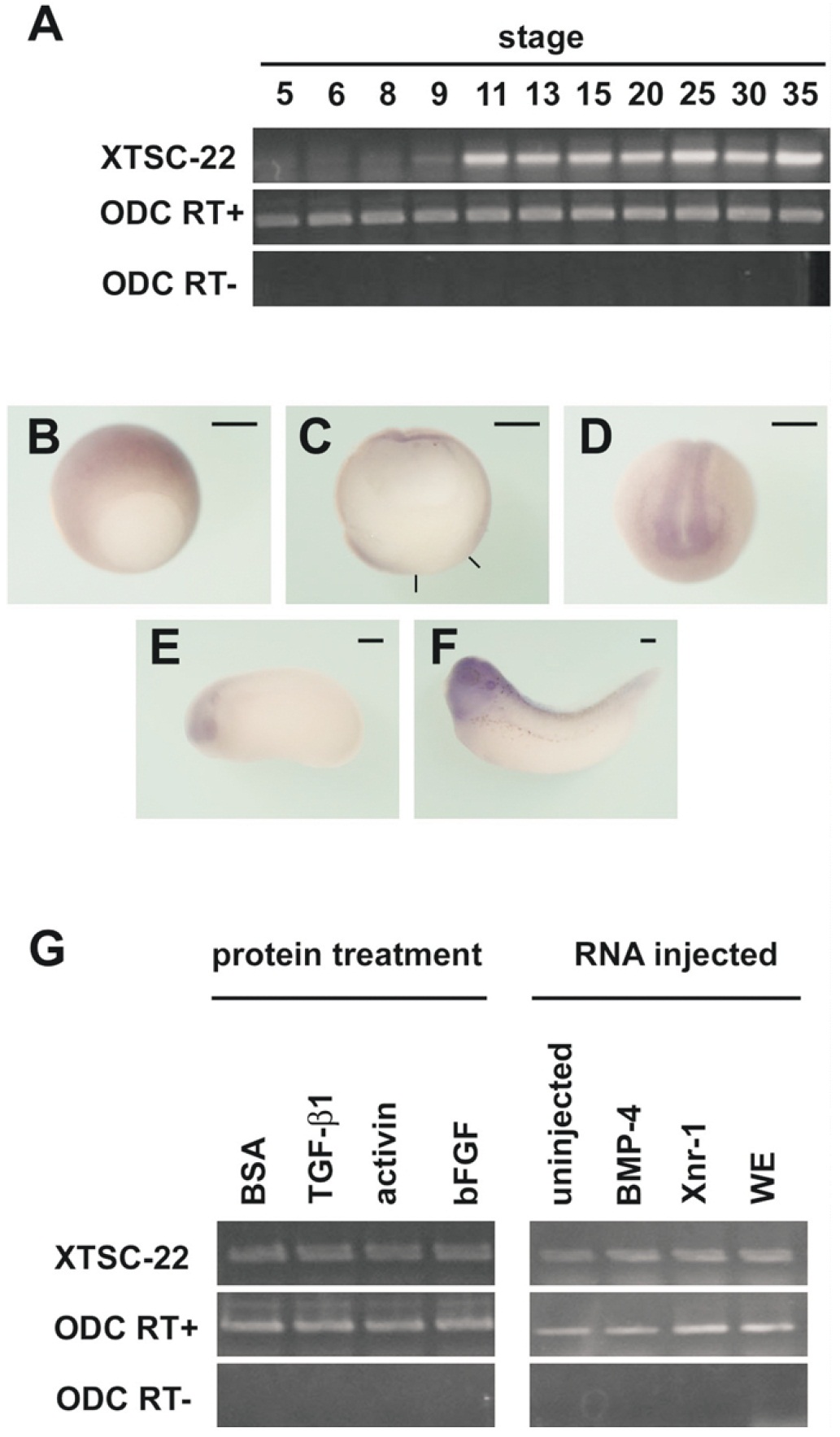
Fig. 2. Expression pattern of TSC-22 (a) Temporal expression pattern of XTSC-22 mRNA detected by reverse transcriptionpolymerase chain reaction (RTPCR). Transcripts were first detected at late blastula (stage 9) and persisted until tadpole stage (stage 35). Ornitine decarboxylase (ODC) expression serves as the quantitative control. RT-PCR was carried out to avoid contamination with genomic DNA. (bâf) Spatial expression pattern of XTSC-22 determined by wholemount in situ hybridization. At stage 10 (b,c), the ectoderm was stained. (c) Sagittal section of the stage-10 embryo in (b). The black lines indicate the blastopore. At neurula stage (d,e), transcripts were observed in the neural region including eyes and brain. This localization was clear at stage 35 (f). Bars, 0.3 mm (g) Induction of XTSC-22 transcription by growth factors: there was no change in XTSC-22 expression with any treatment. WE, whole embryo.
Image published in: Hashiguchi A et al. (2004)
Copyright © 2004. Image reproduced with permission of the Publisher, John Wiley & Sons.
| Gene | Synonyms | Species | Stage(s) | Tissue |
|---|---|---|---|---|
| tsc22d1.L | LOC108709791, tsc-22, tsc22 | X. laevis | Throughout NF stage 18 | neural plate pre-chordal neural plate chordal neural plate anterior neural fold |
| tsc22d1.L | LOC108709791, tsc-22, tsc22 | X. laevis | Throughout NF stage 24 | otic placode optic vesicle brain forebrain midbrain hindbrain |
| tsc22d1.L | LOC108709791, tsc-22, tsc22 | X. laevis | Throughout NF stage 32 | central nervous system brain eye otic vesicle forebrain midbrain hindbrain |
| tsc22d1.L | LOC108709791, tsc-22, tsc22 | X. laevis | Sometime during NF stage 9 to NF stage 11 | ectoderm |
Image source: Published
Permanent Image Page
Printer Friendly View
XB-IMG-133143