XB-IMG-124791
Xenbase Image ID: 124791
|
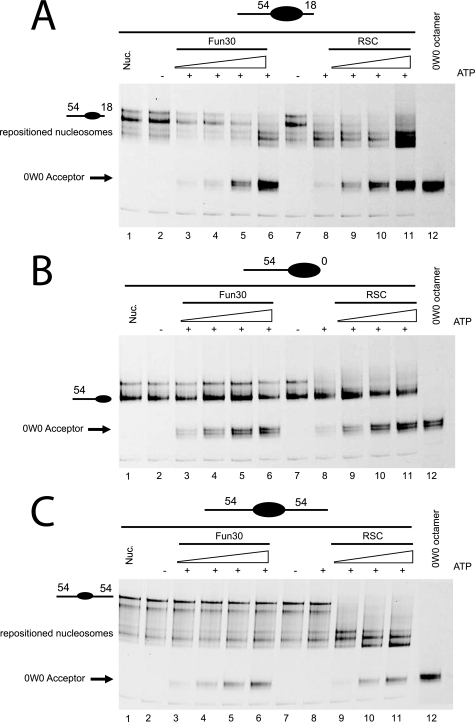
FIGURE 6. Fun30 has higher activity in histone dimer exchange than nucleosome repositioning. 10-μl reactions containing 0.25 pmol of nucleosomes (250 nm) in which H2B is fluorescently labeled with Cy5 assembled at the murine mammary tumor virus nucleosome A (NucA) positioning sequence flanked by the indicated lengths of linker DNA were incubated with Fun30 (100, 200, 300, and 400 pm, lanes 3–6) or RSC (100, 200, 300, and 400 pm, lanes 8–11) in the presence of 0.75 pmol of histone tetramers assembled onto 147-bp DNA (0W0). In each panel, lane 1 contains nucleosomes assembled on the appropriate donor DNA fragment and lane 12 contains the 0W0 fragment assembled with an octamer including fluorescently labeled H2B. Following native gel electrophoresis, the fate of H2B was monitored by fluorescent scanning of the gels. In some cases the signal moves to a location consistent with repositioning of nucleosomes on the donor DNA fragment. In others, transfer to the 0W0 acceptor DNA, which has a distinct mobility, could be detected. Fun30 was observed to cause dimer exchange even in circumstances where repositioning was inefficient. A, the donor nucleosome has an asymmetric linker DNA of 54 bp on one end and 18 bp on the other (54A18). B, the donor nucleosome has a 54-bp linker DNA on one end and 0 bp on the other (54A0). C, the donor nucleosome has 54-bp linkers on either side (54A54). Image published in: Awad S et al. (2010) © 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
