XB-IMG-127384
Xenbase Image ID: 127384
|
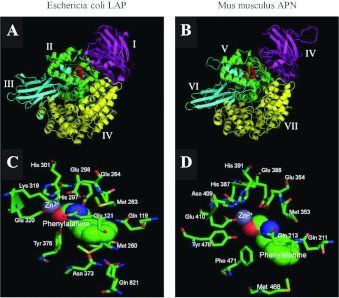
Figure 7. Homology model of mouse APNThe homology model of mouse APN was generated using HHpred Homology detection and structure prediction tool (Max-Planck Institute for Developmental Biology; http://toolkit.tuebingen.mpg.de/hhpred) and visualized using PyMOL v9.9 (DeLano Scientific; http://www.pymol.org). Depicted are E. coli LAP (PDB code 3B34) (A) and murine APN (B) with phenylalanine bound to their active site. The top panels display a lateral three-dimensional view of the murine and E. coli structures. Domains are shaded as follows: magenta (domain I for E. coli, domain IV for mouse); green (domain II for E. coli, domain V for mouse); cyan (domain III for E. coli, domain VI for mouse); and yellow (domain IV for E. coli, domain VII for mouse). Domains I–III from mouse APN are not depicted (residues 1 to 89) as no homologous sequence exists in E. coli for these. The HEXXH……….BXLXE and GXMEN consensus sequences for zinc and amino acid binding are shaded red and orange respectively. The zinc ion is shaded blue-grey. The active sites of E. coli LAP and murine APN are depicted in panels (C) and (D) respectively. The zinc ion is shaded blue-grey, and the bound phenylalanine is depicted as a space-filled model, with oxygen and nitrogen atoms coloured red and blue respectively. All residues predicted to bind and interact with the substrate are shown and labelled. Image published in: Fairweather SJ et al. (2012) © 2012 The Author(s) The author(s) has paid for this article to be freely available under the terms of the Creative Commons Attribution Non-Commercial Licence (http://creativecommons. Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
