XB-IMG-125347
Xenbase Image ID: 125347
|
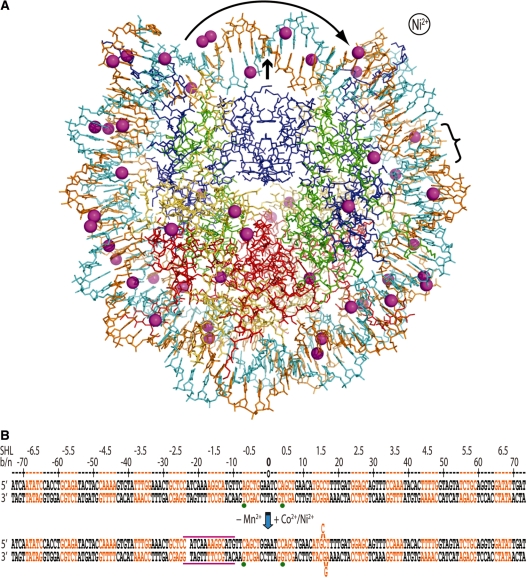
Figure 1. The presence of Co2+ or Ni2+ induces a shift in histone–DNA register and distortions in the double helix. (A) The Ni2+-NCP structure viewed along the particle pseudo 2-fold axis (small arrow). The 47 Ni2+ ions appear as magenta spheres, the two DNA strands are orange and cyan and histone proteins are shown in blue (H3), green (H4), yellow (H2A) and red (H2B). Ni2+ or Co2+ association causes displacement of a bp from one particle half to the opposing half (curved arrow; bracket indicates location of base flip-out distortion). This is also apparent from asymmetry in metal binding about the dyad axis. (B) Minor and major groove-inward sections of NCP147 are orange and black, respectively. The base or nucleotide numbering scheme (b/n) is relative to Mn2+-NCP (top) and corresponds to the 5′ (−) to 3′ (+) direction of either DNA strand in the duplex (SHL = superhelix location, turns from center). A gap in the sequence represents a shift in histone-bp register from DNA stretching (approximate expanse of distortion, magenta lines), which is accompanied by base expulsion at the 1.5-turn location in Co2+-NCP and Ni2+-NCP. Green dots represent two strong peaks in the anomalous difference map that correspond to Co2+/Ni2+-guanine coordination in the refined model (bottom), whereas they coincide with inappropriate binding sites in the Mn2+-NCP model (top). Image published in: Mohideen K et al. (2010) © The Author(s) 2010. Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
