XB-IMG-125175
Xenbase Image ID: 125175
|
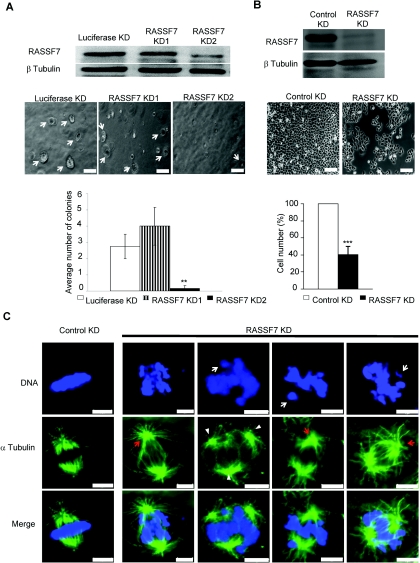
Figure 2. RASSF7 knockdown caused a reduction in cell number and defects in mitosis(A) RASSF7 siRNA oligonucleotides KD2, but not KD1, blocked RASSF7 expression and prevented H1792 cells from forming colonies (arrows). Quantification is based on the number of colonies bigger than 50 μm (**P<0.01 for RASSF7 KD2 compared with Luciferase KD and RASSF7 KD1). (B) shRNA knockdown of RASSF7 reduced HeLa cell numbers. The difference between control and RASSF7-depleted cells was 2.6±0.6-fold (***P<0.001), based on five independent experiments. This was not due to variations in the selection process as it was observed when cells were selected, replated at the same density and then allowed to grow (1.9±0.4-fold reduction). (C) RASSF7-knockdown HeLa cells exhibit defects in spindle formation (red arrows highlight the more radial microtubules) and chromosomal congression, with an increase in cells which had failed to align their DNA (63% compared with 18% in controls, n=300, P<0.01) and an increase in cells with lagging chromosomes (21.5% compared with 6% in controls, n=300, P<0.01, white arrows). There was also a small increase in tripolar spindles (32.6% compared with 23.7% in controls, n=300, P<0.05, arrowheads). Scale bars, 100 μm (A and B) and 5 μm (C). Image published in: Recino A et al. (2010) © 2010 The Author(s) The author(s) has paid for this article to be freely available under the terms of the Creative Commons Attribution Non-Commercial Licence (http://creativecommons. Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
