XB-IMG-127582
Xenbase Image ID: 127582
|
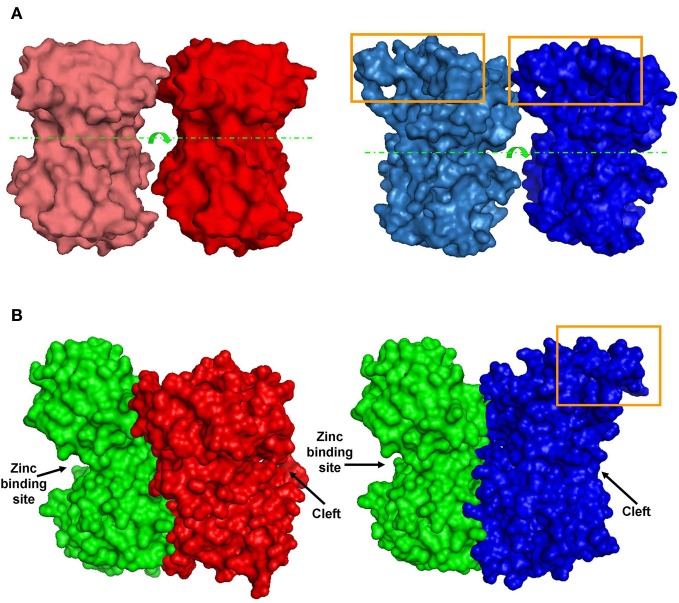
Figure 6. Predicted dimer structures. (A) GluN1 homodimers. Left (red)—A 3D model of GluN1-1a↔GluN1-1a NTD interactions. Right (blue)—A 3D model of GluN1-1b↔GluN1-1b NTD interactions (manual docking). Box—the extra 21 amino–acid-loop (exon 5). This structure might interfere with the interaction of GluN1-1b subunits. Broken lines indicate the cleft. Arrows represent the cleft's direction. (B) GluN1-GluN2 heterodimer 3D structures. Left—docking of GluN1-1a NTD (red) to GluN2A NTD (green). Right—docking of GluN1-1b NTD (blue) to GluN2A NTD (green). Note that the loop of 21 extra amino acids in GluN1-1b (orange box) faces out and does not interfere with sub-domain interaction. The figure was created using PyMOL. Image published in: Mor A et al. (2012) Copyright © 2012 Mor, Kuttner, Levy, Mor, Hollmann and Grossman. Creative Commons Attribution license Larger Image Printer Friendly View |
