XB-IMG-127342
Xenbase Image ID: 127342
|
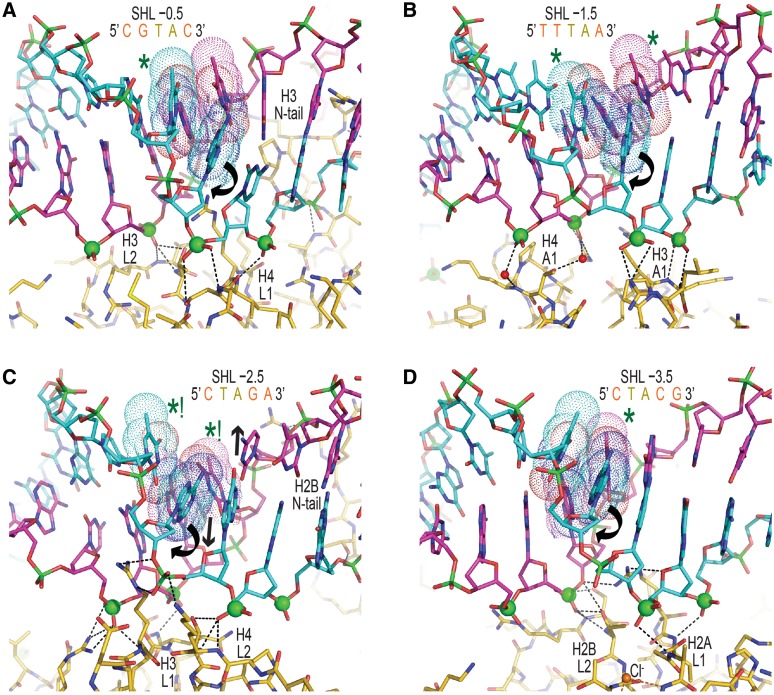
Figure 4. DNA binding and structure over TA elements in NCP-601L. (A–D) TA dinucleotides (bases, space filling dots) are situated at the pressure points of the centrally located histone binding sites, where they display a diversity in conformational distortions. Rotational dislocation of bases within the TA step (curved arrows, only front strand shown for clarity) permits unabated compression of the minor groove for fitting of the binding platform (phosphorous atoms, spheres) to the histone surface. DNA–histone hydrogen bonds appear as black dashed lines. Permanganate reactivity hotspots are designated with asterisks, whereby that at SHL ±2.5 is by far the most prominent as a consequence of extreme base unstacking promoted by base pair displacement into the minor and major grooves via shift (C, arrows). Note that shift is also the primary DNA structural parameter influencing platinum drug reaction, since it dictates solvent access to the major groove edge (14). Image published in: Chua EY et al. (2012) © The Author(s) 2012. Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
