XB-IMG-123858
Xenbase Image ID: 123858
|
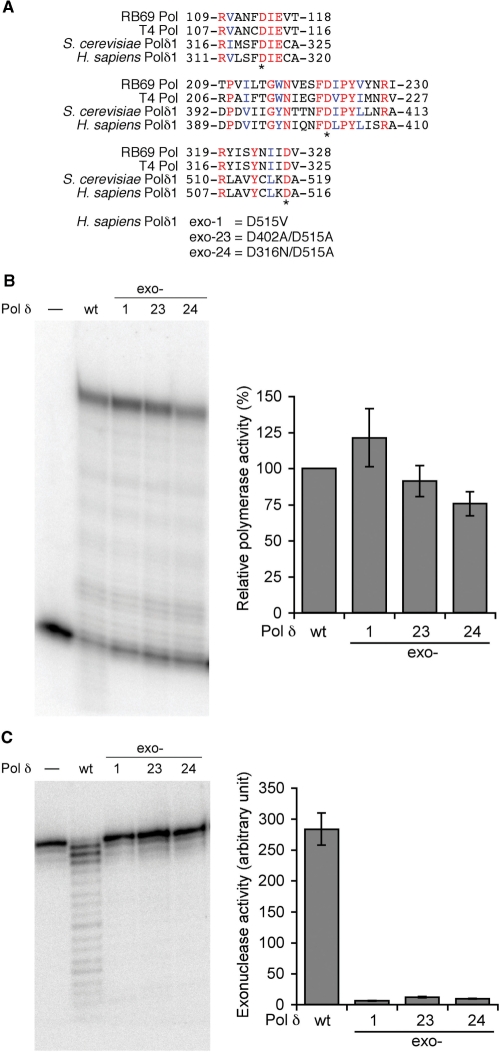
Figure 5. Site-directed mutants of Pol δ exonuclease domains. (A) Amino-acid sequence comparison of the exonucelase domains of replicative DNA polymerases. Budding yeast and human Pol δ catalytic subunits are compared to DNA polymerases from bacterophages, RB69 and T4. Identical residues and similar residues conserved among the four enzymes are indicated in red and blue fonts, respectively. Three aspartic acids presumed to be critical for exonuclease activity are indicated with asterisks. Mutations introduced into human Pol δ in this study are indicated below. (B) DNA polymerase activity with wild-type and three mutant human Pol δ enzymes. The assay was conducted with the normal primer/49-nt G templates in the presence of PCNA, RFC and the Lac repressor as described in ‘Materials and Methods’ section. The activity was calculated as [intensity of the fully extended product]/([intensity of the fully extended product] + [intensity of the unextended primer]), and normalized with the activity of the wild-type enzyme as 100%. Average activities and standard deviations of mutant enzymes from three experiments were presented in the right panel. (C) Exonuclease assay of Pol δ. 5′-Labeled single-stranded oligonucleotide (ssDNA primer in Figure 2) was incubated with indicated Pol δ enzymes. The exonuclease activity was calculated as described in ‘Materials and Methods’ section. Average activities and standard deviations from three reactions were presented in the right panel. Image published in: Fazlieva R et al. (2009) © 2009 The Author(s). Creative Commons Attribution-NonCommercial license Larger Image Printer Friendly View |
