XB-IMG-126588
Xenbase Image ID: 126588
|
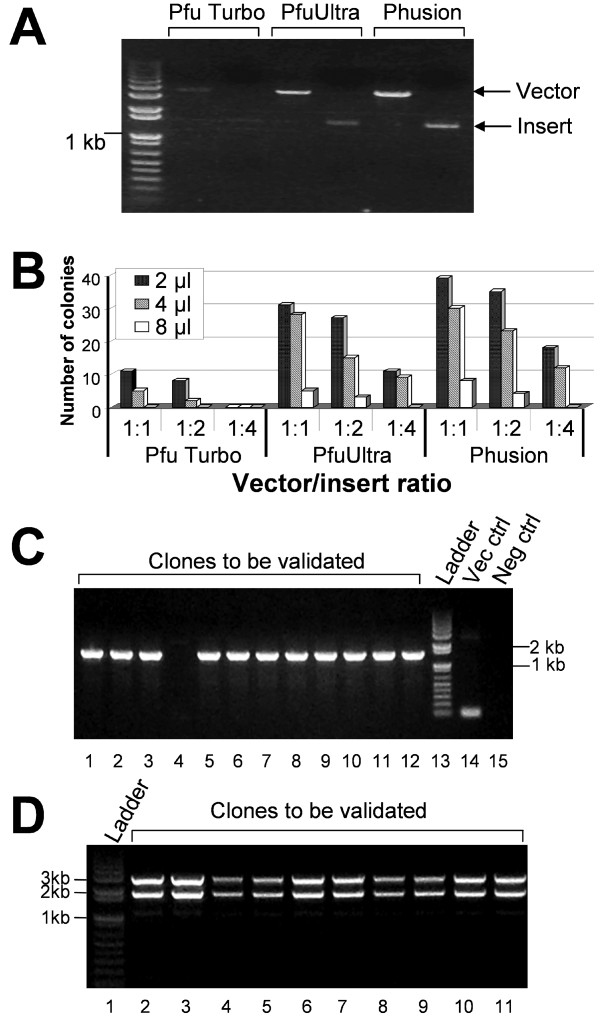
Figure 4. Optimization of cloning conditions. (A) PCR amplification of a target cDNA (human nAChR α9 subunit) and the pGEMHE vector using different DNA polymerases: Pfu Turbo, PfuUltra and Phusion. (B) Comparison of number of colonies grown on the plates after transformation. Three different vector-to-insert ratios (1:1, 1:2, and 1:4) during DpnI digestion and three amounts of vector-insert mixtures (2, 4, and 8 μl) for transformation were tested. See text for details. (C) Clone validation by PCR using GoTaq DNA polymerase. Lanes 1 to 12: target clones to be validated; Lane 13: 1 Kb plus DNA ladder; Lane 14: pGEMHE vector control; Lane 15: negative control using pCR4-TOPO-α9 parent plasmid. (D) Clone validation by restriction digestion to exclude unusual constructs. Lane 1: 1 Kb plus DNA ladder, Lanes 2-11: target clones double digested with KpnI and NheI. Note that this digestion resulted in a pGEM vector and an insert with α9 nAChR plus the 5'UTR and 3'UTR of Xenopus β-globin. Image published in: Li C et al. (2011) Copyright ©2011 Li et al; licensee BioMed Central Ltd. Creative Commons Attribution license Larger Image Printer Friendly View |
