XB-IMG-159752
Xenbase Image ID: 159752
|
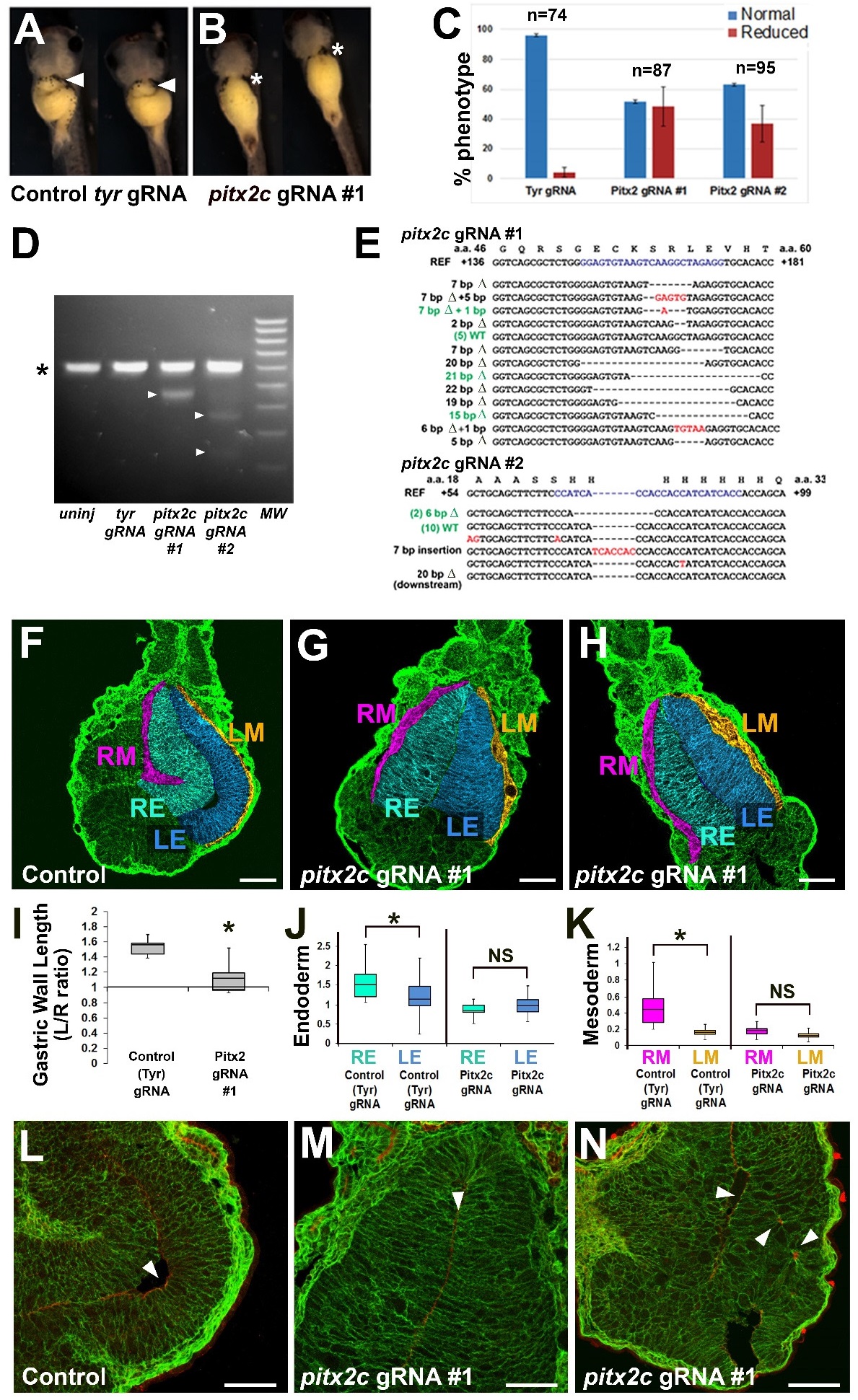
Figure S5. CRISPR-Cas9 mediated editing of pitx2c gene perturbs stomach asymmetry. Xenopus embryos
were injected with 2 ng synthetic Cas9 mRNA plus 300 pg control tyrosinase (tyr) gRNA (A) or pitx2c gRNA
(B), and allowed to develop until stage 42. The graph (C) indicates the percentage of embryos in which the
greater curvature of the stomach is normal (arrowhead in A) or reduced/absent (* in B) after injection with the
tyr gRNA or two different pitx2c gRNAs (#1 and #2). D) Genomic DNA from 10 embryos injected with each
gRNA was pooled and PCR amplified using exon 1-specific primers, and then tested for CRISPR/Cas9-induced
mutations by T7 endonuclease I assay. The asterisk (*) indicates the 500 bp amplicon not cut in un-injected or
tyr gRNA-injected control embryos. Arrowheads indicate bands resulting from mismatches (inferred mutations)
in amplicons cleaved by T7 endonuclease I. E) Sequencing of a subset of individual clones validates the
presence of deleterious mutations in pitx2c gRNA injected embryos. For pitx2c gRNA #1, 9/17 mutations were
likely nulls and 3/17 were predicted to result in compromised function; for pitx2c gRNA #2: 2/18 mutations
were likely nulls and 4/17 were likely to result in compromised function. F-N) Sections through stomachs of
Cas9 control and pitx2c gRNA (#1) injected embryos (Stage 39) were stained for integrin (green) and false
color-coded as in Fig. 1 to highlight the relevant tissue layers (RE, right endoderm; LE, left endoderm; RM,
right mesoderm; LM, left mesoderm). Controls show normal asymmetric expansion of the left stomach wall
(F), but this is eliminated in embryos injected with pitx2c gRNA (G-H). Normal asymmetries in the lengths of
the left and right stomach walls (I), and the widths of the endoderm (Endo) and mesoderm (Meso) layers (J-K),
are also significantly perturbed by CRISPR-Cas editing of pitx2c; * denotes p<0.01. Compared to controls (L),
left endoderm tissue architecture is severely disrupted, and apicobasal polarity is reduced (M), and/or
disorganized (N) in embryos with CRISPR-mediated mutations in pitx2c; arrowheads indicate the expression of
the apical marker aPKC (red). Scalebars= 100 ïM. Image published in: Davis A et al. (2017) Copyright © 2017. Image reproduced with permission of the Publisher and the copyright holder. This is an Open Access article distributed under the terms of the Creative Commons Attribution License.
Image source: Published
Larger Image Printer Friendly View |
