XB-IMG-195561
Xenbase Image ID: 195561
|
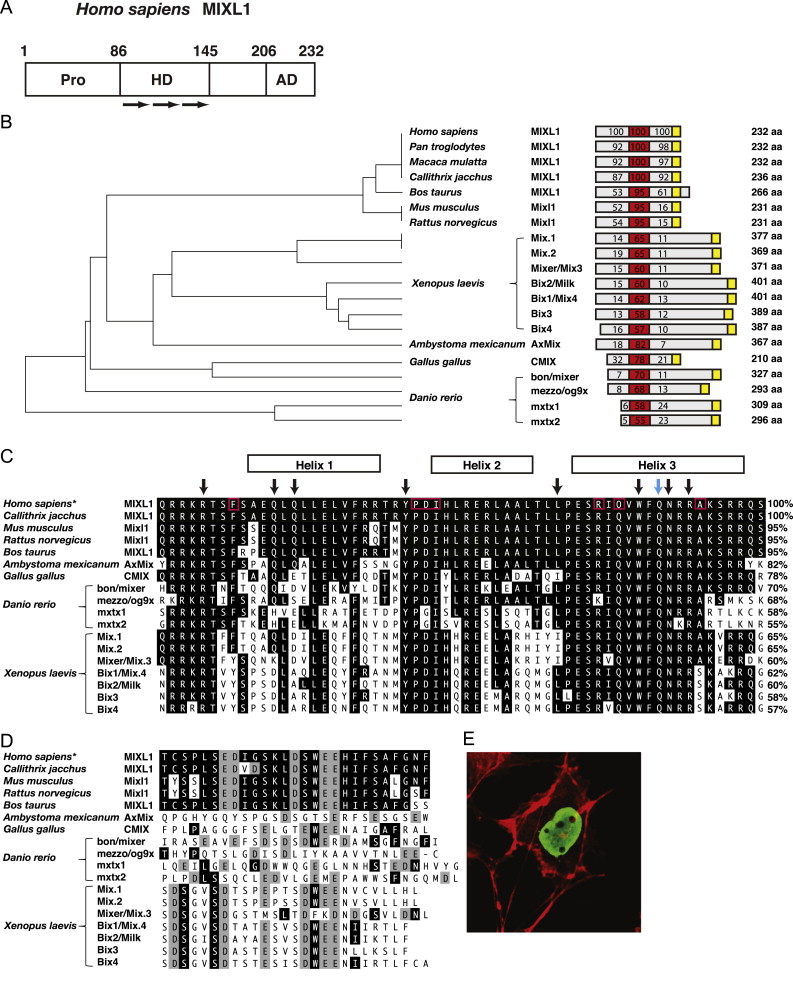
Fig. 1. Domain organisation of Mix/Bix proteins. (A) As with the other Mix/Bix family members, human MIXL1 can be divided into functional regions as indicated. The amino-terminal region contains a number of conserved proline (Pro) residues, whilst the central region incorporates the DNA binding homeodomain (HD) composed of three alpha helices (arrows). The carboxy-terminal region contains the signature residues of an acidic transcriptional activation domain (AD). (B) Phylogenetic analysis of Mix/Bix proteins. For each protein a scaled cartoon depicts its relative size. The total amino acid length is listed on the right. The percentage identity, when aligned with human MIXL1, is shown for the homeodomain (red), sequence N-terminal to the homeodomain (grey) and sequence C-terminal to the homeodomain (grey and yellow). The position of the activation domain is shown in yellow. (C) An alignment of the human MIXL1 homeodomain amino acid sequence with other Mix-related proteins. Residues that match with human MIXL1 have been shaded black. The percentage identities are listed on the right. The location of the three predicted alpha helices is shown above. The black arrows denote eight amino acids characteristic of homeodomain proteins. The amino acids boxed in red are shared between most Mix family genes and the paired homeodomain sequence but generally are not found in other homeodomains (Rosa, 1989), the blue arrow denotes the conserved glutamine at position 50 (Q50) that influences DNA sequence binding specificity (Wilson et al., 1993). *The Homo sapiens homeodomain sequence shown is identical to that of Pan troglodytes and Macaca mulatta. (D) A comparison of Mix family carboxy-termimal amino acid sequences. Acidic residues have been shaded grey. *The Homo sapiens carboxy-termimal sequence shown is identical to that of Pan troglodytes and Macaca mulatta. (E) Nuclear localisation of Mixl1 protein. Immunofluorescent image of COS-7 cells transfected with an expression vector encoding mouse Mixl1 stained with anti-Mixl1 monoclonal antibody generated to a carboxy terminal (6G2) peptide. Anti-Mixl1 staining (Green) was combined with the detection of cytoplasmic actin with phalloidin-TRITC (Red). Panels C and D are adapted from research originally published in Development Dynamics. Robb, L., Hartley, L., Begley, C.G., Brodnicki, T.C., Copeland, N.G., Gilbert, D.J., Jenkins, N.A. and Elefanty, A.G. Cloning, expression analysis, and chromosomal localization of murine and human homologues of a Xenopus gene. Development Dynamics. 2000; 219:497–504. Copyright © 2000 Wiley-Liss, Inc. Panel E is from research originally published in Stem Cells and Development. Mossman, A.K., Sourris, K., Ng, E., Stanley, E.G. and Elefanty, A.G. Mixl1 and Oct4 proteins are transiently co-expressed in differentiating mouse and human embryonic stem cells. Stem Cells and Development. 2005;14:656-663. Copyright © 2011, Mary Ann Liebert, Inc. publishers. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) Image published in: Pereira LA et al. (2012) Copyright © 2012. Image reproduced with permission of the Publisher, Elsevier B. V. Larger Image Printer Friendly View |
