XB-IMG-118819
Xenbase Image ID: 118819
|
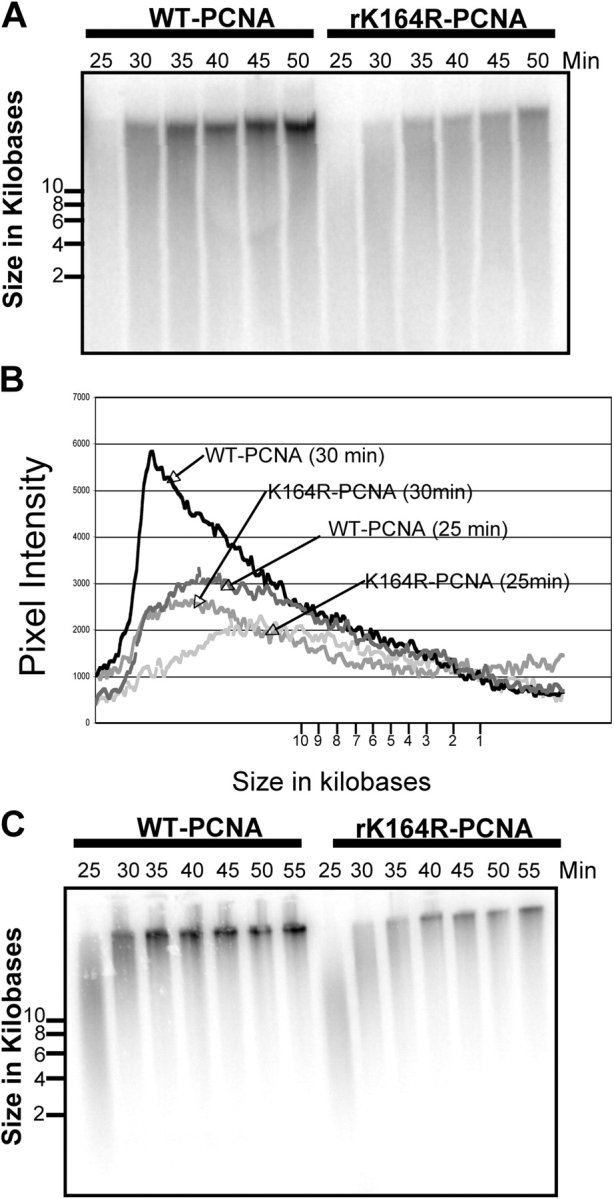
Figure 6. Loss of PCNA ubiquitylation slows replication fork progression. (A) Sperm chromatin was incubated in X. laevis egg extract containing either 0.2 μg/μL PCNA (wild type) or 0.2 μg/μL PCNA (K164R). Aliquots were removed after 25, 30, 35, 40, 45, and 50 min and prepared for alkaline agarose gel analysis as described in Materials and methods. (B) Fragment lengths were calculated by comparison to a DNA ladder and plotted against pixel intensity obtained with National Institutes of Health Image software. (C) Sperm chromatin was incubated in X. laevis egg extract containing either 0.2 μg/μL PCNA (wild type) or 0.2 μg/μL PCNA (K164R). Cold dATP was added after 25 min. Aliquots were removed after 25, 30, 35, 40, 45, and 50 min and prepared for alkaline agarose gel analysis as described in Materials and methods. Image published in: Leach CA and Michael WM (2005) Copyright © 2005, The Rockefeller University Press. Creative Commons Attribution-NonCommercial-ShareAlike license Larger Image Printer Friendly View |
