Morpholino Side Effects, cont.
Morpholinos Do Not Elicit an Innate Immune Response during Early Xenopus Embryogenesis
Kitt D. Paraiso, Ira L. Blitz, Jeff J. Zhou, Ken W.Y. Cho
Developmental Cell, Volume 49, Issue 4, 20 May 2019, Pages 643-650.e3
Click here to view article at Developmental Cell.
Click here to view article on Pubmed.
Click here to view article on Xenbase.
Highlights
Analyzed publicly available Xenopus morphant RNA-seq datasets
Innate immune response gene induction is not a general effect related to morpholinos
Strong induction of an immune response is specific to the tbxt/tbxt2 morpholinos
* this article is in response to Gentsch et al. (2018) as reported on Xenbase, Pubmed, and Developmental Cell
Abstract
It has recently been reported that a common side effect of translation-blocking morpholino antisense oligonucleotides is the induction of a set of innate im- mune response genes in Xenopus embryos and that splicing-blocking morpholinos lead to unexpected off-target mis-splicing events. Here, we present an analysis of all publicly available Xenopus RNA sequencing (RNA-seq) data in a reexamination of the effects of translation-blocking morpholinos on the innate immune response. Our analysis does not support the authors’ general conclusion, which was based on a limited number of RNA-seq datasets. Moreover, the strong induction of an immune response appears to be specific to the tbxt/tbxt2 morpholinos. The more comprehensive study pre- sented here indicates that using morpholinos for targeted gene knockdowns remains of considerable value for the rapid identification of gene function.
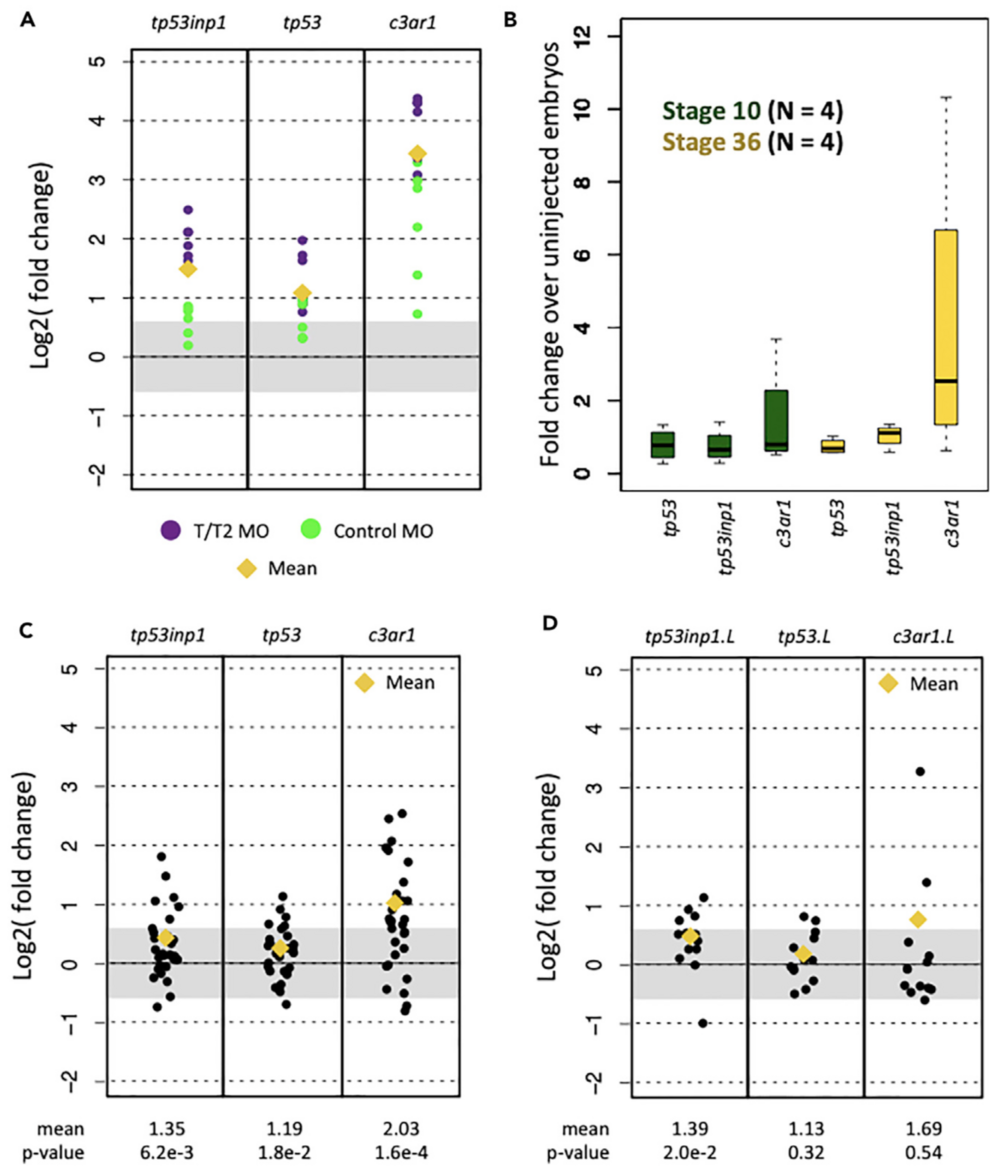
Figure 1. Expression of Innate Immune Response Genes in X. tropicalis and X. laevis RNA-Seq Datasets
(A) Fold change in induction caused by the tbxt/ tbxt2 MOs and the control MOs.
(B) Fold change caused by control MO in biological replicates at stages 10 and 36 using RT-qPCR.
(C and D) Fold-change induction of innate immune response genes in X. tropicalis (C) and X. laevis (D) datasets.
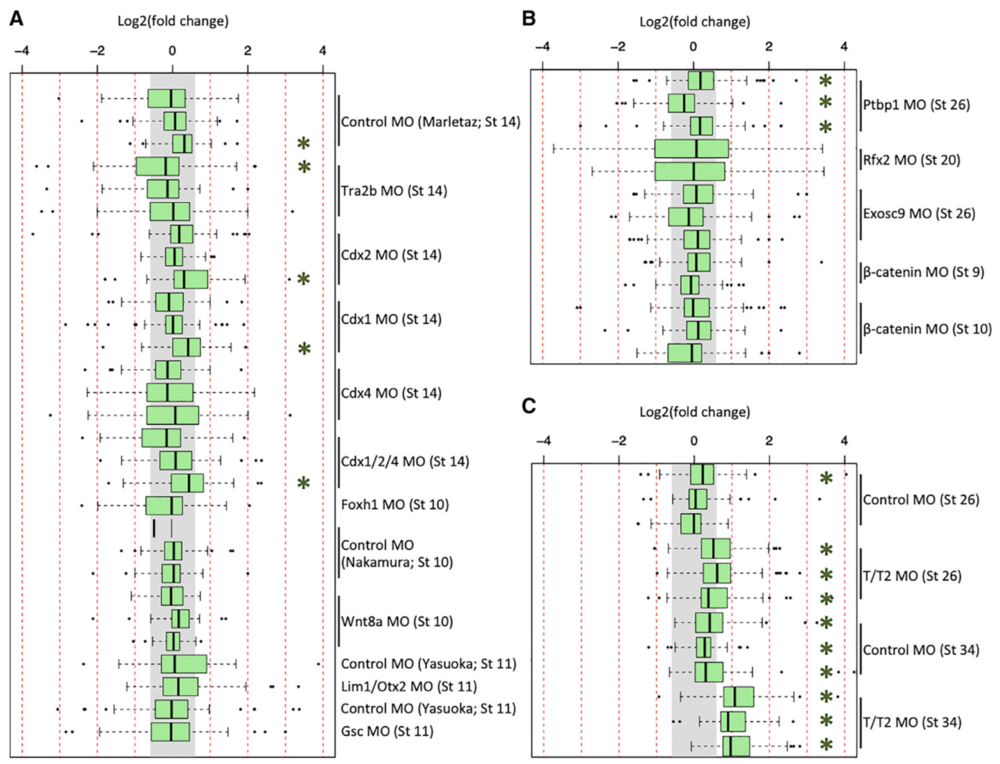
Figure 2. Expression of GO-Identified Innate Immune Response Genes in X. tropicalis and X. laevis RNA-Seq Datasets
(A–C) Fold-change expression of innate immune response genes across 29 datasets in X. tropicalis (A), across 13 datasets in X. laevis (B), and in 12 of the tbxt/t2 MO datasets (C). The gray region indicates fold change of <1.53. Green asterisk (*) indicates a t test p value of <0.01.
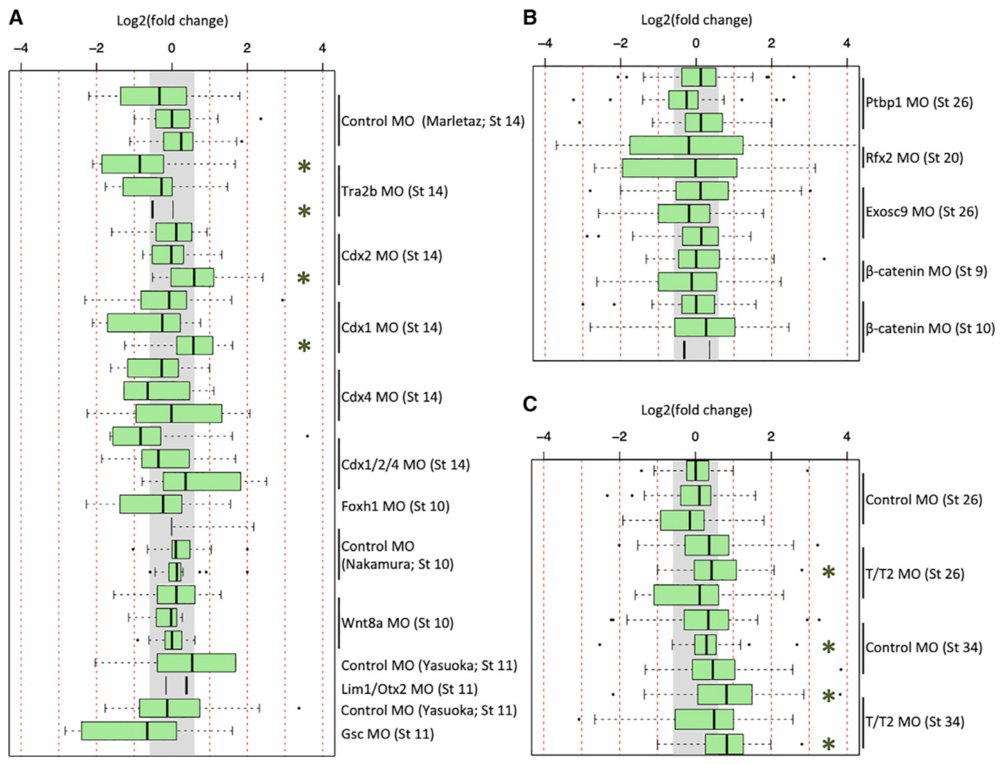
Figure 3. Expression of Literature-Identified Innate Immune Response Genes in X. tropicalis and X. laevis RNA-Seq Datasets
(A–C) Fold-change expression of innate immune response genes across 29 datasets in X. tropicalis (A), 13 datasets in X. laevis (B), and in 12 of the tbxt/t2 MO datasets (C). Gray region indicates fold change of < 1.53. Green asterisk (*) indicates a t test p value of < 0.01.
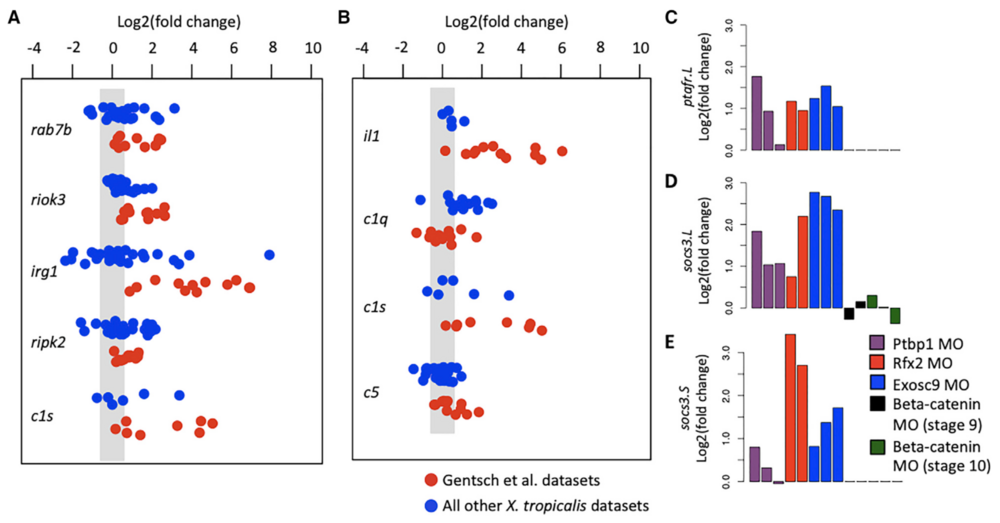
Figure 4. Specific Induction of Innate Immune Response Genes
(A and B) Fold-change expression of genes, which were identified to be significantly activated in the X. tropicalis datasets in both the GO-identified (A) and the literature-identified (B) cohort of genes.
(C–E) Fold-change expression of X. laevis genes ptafr.L/gene13059 (C), socs3.L/gene3766 (D), and socs3.S/gene50103 (E), which were identified to be signif- icantly activated. We used the criteria p value < 0.01 and fold change > 1.5 to define significant.
Adapted with permission from Elsevier on behalf of Developmental Cell: Paraiso et al., Morpholinos Do Not Elicit an Innate Immune Response during Early Xenopus Embryogenesis. Developmental Cell 49, 643–650 May 20, 2019. Copyright 2019 Elsevier Inc. https://doi.org/10.1016/j.devcel.2019.04.019
