CRISPR/Cas9-based simple transgenesis in Xenopus laevis
Dev Biol 2022 Sep 01;489:76-83. doi: 10.1016/j.ydbio.2022.06.001.
Shibata Y, Suzuki M, Hirose N, Takayama A, Sanbo C, Inoue T, Umesono Y, Agata K, Ueno N, Suzuki KT, Mochii M.
Click here to view article at Developmental Biology
Click here to view article on PubMed.
Click here to view article on Xenbase.
Abstract
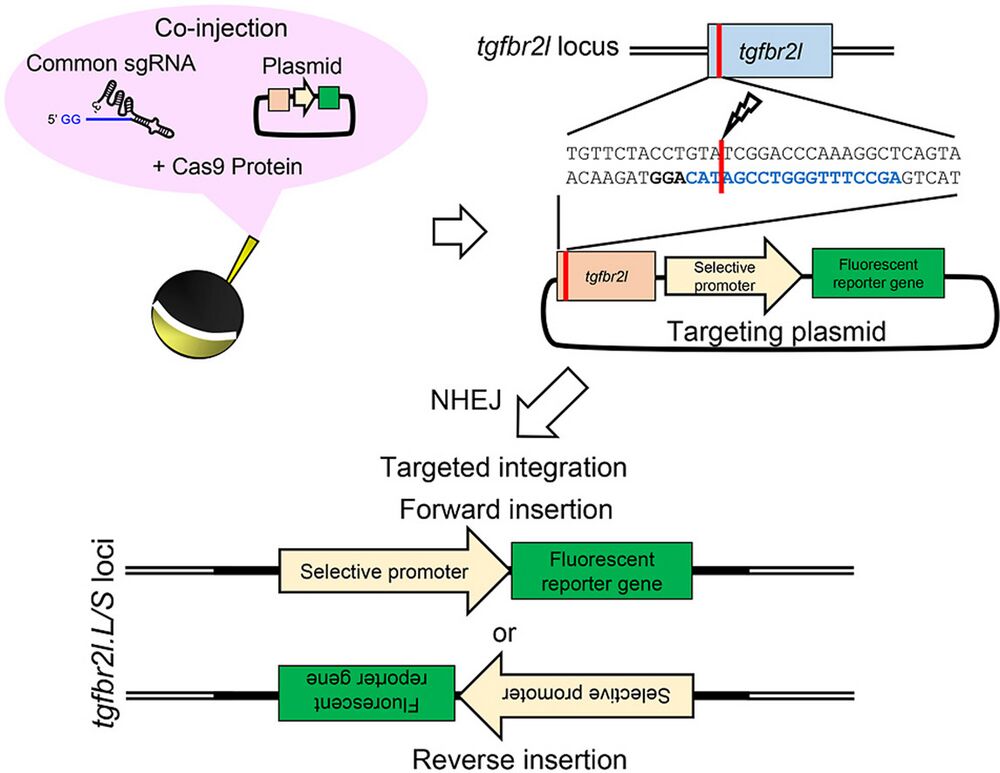
Transgenic techniques have greatly increased our understanding of the transcriptional regulation of target genes through live reporter imaging, as well as the spatiotemporal function of a gene using loss- and gain-of-function constructs. In Xenopus species, two well-established transgenic methods, restriction enzyme-mediated integration and I-SceI meganuclease-mediated transgenesis, have been used to generate transgenic animals. However, donor plasmids are randomly integrated into the Xenopus genome in both methods. Here, we established a new and simple targeted transgenesis technique based on CRISPR/Cas9 in Xenopus laevis. In this method, Cas9 ribonucleoprotein (RNP) targeting a putative harbor site (the transforming growth factor beta receptor 2-like (tgfbr2l) locus) and a preset donor plasmid DNA were co-injected into the one-cell stage embryos of X. laevis. Approximately 10% of faithful reporter expression was detected in F0 crispants in a promoter/enhancer-specific manner. Importantly, efficient germline transmission and stable transgene expression were observed in the F1 offspring. The simplicity of this method only required preparation of a donor vector containing the tgfbr2l genome fragment and Cas9 RNP targeting this site, which are common experimental procedures used in Xenopus laboratories. Our improved technique allows the simple generation of transgenic X. laevis, so is expected to become a powerful tool for reporter assay and gene function analysis.
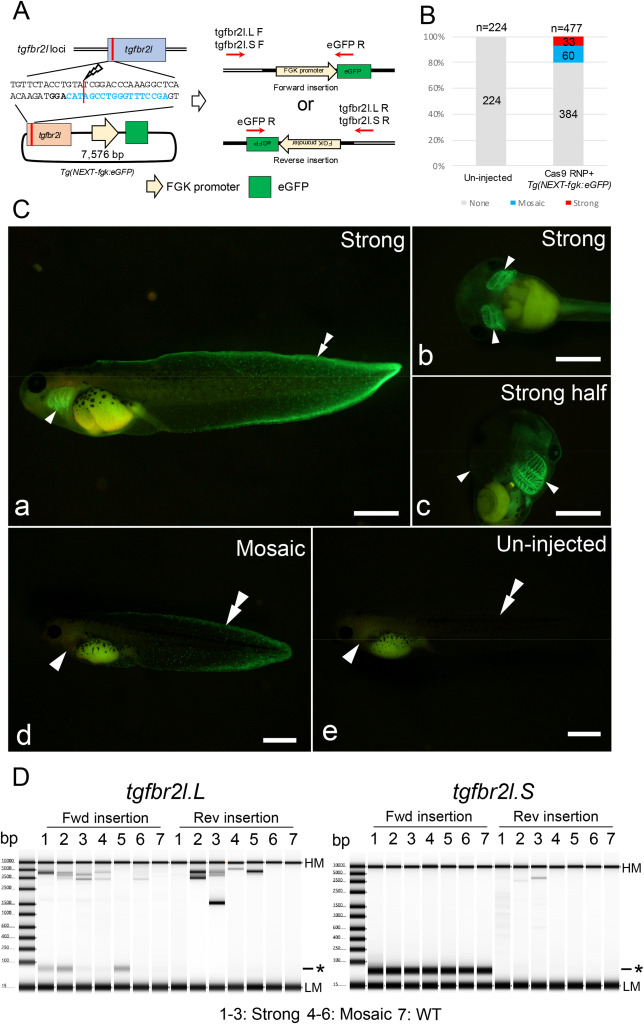
Fig. 1. Generation of transgenic Xenopus with FGK promoter-dependent eGFP expression. (A) Schematic of Tg(NEXT-fgk:eGFP) integration at the tgfbr2l locus. (B) Phenotypes were classified into three groups depending on the fluorescent intensity and the localization: strong eGFP signal in the fin and gill including half transgenic animals; mosaic, weak eGFP signal only in the fin or gill; and none, no eGFP signals detected. Total and group sample numbers are indicated at the top and middle of each column, respectively. (C) Representative photographs of eGFP signals in Xla.Tg(NEXT-fgk:eGFP). Strong eGFP signals were detected in the fin and gill (a–c); weak eGFP signals were observed at the edge of the fin area (d); while no eGFP signals were detected in un-injected tadpoles (e). Arrowheads and double arrowheads indicate the gill and the edge of the tail fin, respectively. Bars = 1 mm. (D) Genotyping for Tg(NEXT-fgk:eGFP) integration in the tgfbr2l locus by PCR. PCR products were detected between 3.5 kb and 5 kb, suggesting Tg(NEXT-fgk:eGFP) was incorporated into the locus. Samples 1–3, strong eGFP signals were detected in the gill and the edge of the tail fin; samples 4–6, weak signals were only detected in the gill or edge of the tail fin; sample 7, un-injected tadpole. Detailed the electropherogram for the PCR products is indicated in Supplementary Fig. 7. HM: High molecular weight marker, LM: Low molecular weight marker and asterisk: primer dimer in TapeStation electropherogram.
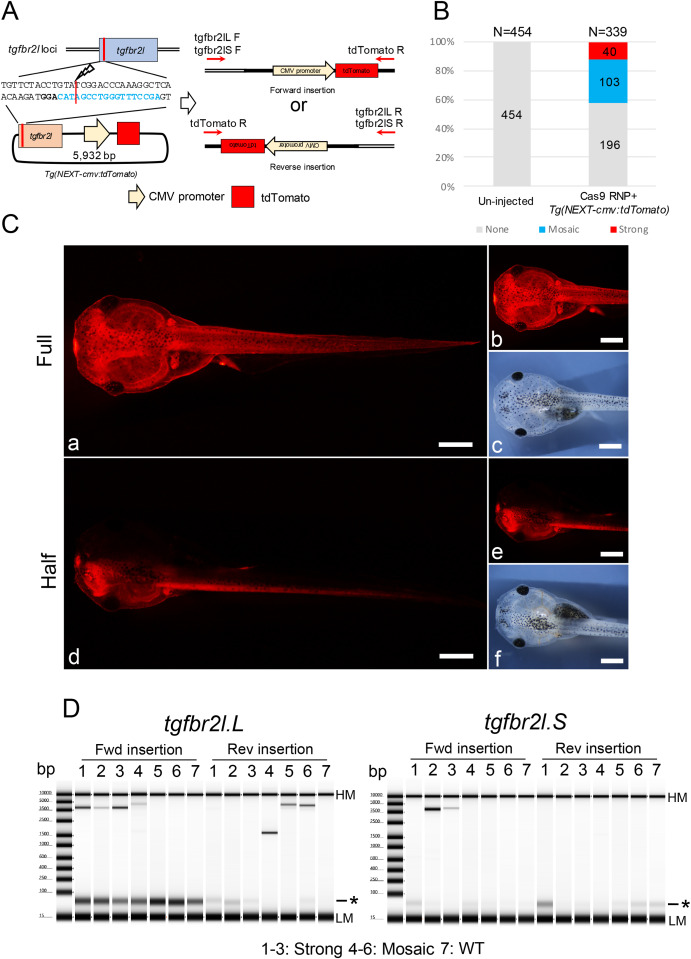
Fig. 2. Generation of transgenic Xenopus with CMV promoter-dependent tdTomato expression. (A) Schematic of Tg(NEXT-cmv:tdTomato) integration at the tgfbr2l locus. (B) Phenotypes were classified into three groups: strong tdTomato signals in the whole body including half transgenic animals; mosaic, weak tdTomato signals in the epithelial cells or muscle cells; and none, no tdTomato signals detected. Total and group sample numbers are indicated at the top and middle of each column, respectively. (C) Representative photographs of tdTomato signals in Xla.Tg(NEXT-cmv:tdTomato). Strong tdTomato signals were detected in the whole body (a, b) including half transgenic animals (d, e). c and f show bright field images of b and e, respectively. Bars = 1 mm. (D) Genotyping for Tg(NEXT-cmv:tdTomato) integration in the tgfbr2l locus by PCR. PCR products were detected between 3.5 kb and 5 kb, suggesting Tg(NEXT-cmv:tdTomato) was incorporated into the locus. Samples 1–3, strong tdTomato signals were detected in the gill and the edge of the tail fin; samples 4–6, weak signals were only detected in the gill or edge of the tail fin; sample 7, un-injected tadpole. Detailed the electropherogram for the PCR products is indicated in Supplementary Fig. 12. HM: High molecular weight marker, LM: Low molecular weight marker and asterisk: primer dimer in TapeStation electropherogram.
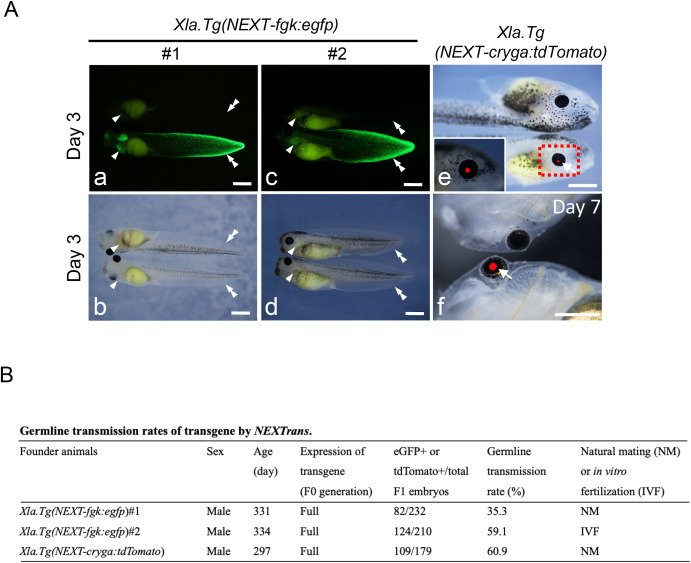
Fig. 4. Germline transmission of various transgenic X. laevis generated by NEXTrans. (A) Representative photographs of F1 offspring in Xla.Tg(NEXT-fgk:egfp) (a–d), and Xla.Tg(NEXT-cryga:tdTomato) (e and f) embryos. Arrowheads and double arrowheads indicate the gill and the fin edge, respectively. b and d show bright field images of a and c, respectively. On the other hand, arrows show the eye with tdTomato signal. Bars = 1 mm. (B) The table summarizes the germline transmission rates of various transgenic animals. F1 embryos were generated by natural mating or in vitro fertilization outcrossed with wild type females. Two adult males were used for Xla.Tg(NEXT-fgk:egfp). The germline transmission rates were scored at stage 40–43.
Adapted with permission from Elsevier/Science Direct on behalf of Developmental Biology: Shibata et al. (2022). CRISPR/Cas9-based simple transgenesis in Xenopus laevis. Dev Biol 2022 Sep 01;489:76-83. doi: 10.1016/j.ydbio.2022.06.001.
Last Updated: 2023-05-23